SARS-CoV-2 Nsp6 damages Drosophila heart and mouse cardiomyocytes through MGA/MAX complex-mediated increased glycolysis
- PMID: 36180527
- PMCID: PMC9523645
- DOI: 10.1038/s42003-022-03986-6
SARS-CoV-2 Nsp6 damages Drosophila heart and mouse cardiomyocytes through MGA/MAX complex-mediated increased glycolysis
Abstract
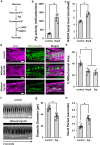
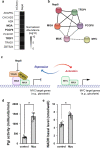
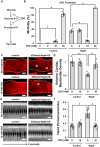
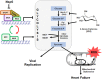
SARS-CoV-2 infection causes COVID-19, a severe acute respiratory disease associated with cardiovascular complications including long-term outcomes. The presence of virus in cardiac tissue of patients with COVID-19 suggests this is a direct, rather than secondary, effect of infection. Here, by expressing individual SARS-CoV-2 proteins in the Drosophila heart, we demonstrate interaction of virus Nsp6 with host proteins of the MGA/MAX complex (MGA, PCGF6 and TFDP1). Complementing transcriptomic data from the fly heart reveal that this interaction blocks the antagonistic MGA/MAX complex, which shifts the balance towards MYC/MAX and activates glycolysis-with similar findings in mouse cardiomyocytes. Further, the Nsp6-induced glycolysis disrupts cardiac mitochondrial function, known to increase reactive oxygen species (ROS) in heart failure; this could explain COVID-19-associated cardiac pathology. Inhibiting the glycolysis pathway by 2-deoxy-D-glucose (2DG) treatment attenuates the Nsp6-induced cardiac phenotype in flies and mice. These findings point to glycolysis as a potential pharmacological target for treating COVID-19-associated heart failure.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
SARS-CoV-2 Nsp6-Omicron causes less damage to the Drosophila heart and mouse cardiomyocytes than ancestral Nsp6.Commun Biol. 2024 Dec 3;7(1):1609. doi: 10.1038/s42003-024-07307-x. Commun Biol. 2024. PMID: 39627475 Free PMC article.
-
SARS-CoV-2 viral genes Nsp6, Nsp8, and M compromise cellular ATP levels to impair survival and function of human pluripotent stem cell-derived cardiomyocytes.Stem Cell Res Ther. 2023 Sep 13;14(1):249. doi: 10.1186/s13287-023-03485-3. Stem Cell Res Ther. 2023. PMID: 37705046 Free PMC article.
-
Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif.EMBO J. 1999 Dec 15;18(24):7019-28. doi: 10.1093/emboj/18.24.7019. EMBO J. 1999. PMID: 10601024 Free PMC article.
-
Interaction of SARS-CoV-2 with cardiomyocytes: Insight into the underlying molecular mechanisms of cardiac injury and pharmacotherapy.Biomed Pharmacother. 2022 Feb;146:112518. doi: 10.1016/j.biopha.2021.112518. Epub 2021 Dec 9. Biomed Pharmacother. 2022. PMID: 34906770 Free PMC article. Review.
-
Myc's secret life without Max.Cell Cycle. 2009 Dec;8(23):3848-53. doi: 10.4161/cc.8.23.10088. Epub 2009 Dec 15. Cell Cycle. 2009. PMID: 19887915 Review.
Cited by
-
SARS-CoV-2 Nsp6-Omicron causes less damage to the Drosophila heart and mouse cardiomyocytes than ancestral Nsp6.Commun Biol. 2024 Dec 3;7(1):1609. doi: 10.1038/s42003-024-07307-x. Commun Biol. 2024. PMID: 39627475 Free PMC article.
-
A comprehensive Drosophila resource to identify key functional interactions between SARS-CoV-2 factors and host proteins.Cell Rep. 2023 Aug 29;42(8):112842. doi: 10.1016/j.celrep.2023.112842. Epub 2023 Jul 20. Cell Rep. 2023. PMID: 37480566 Free PMC article.
-
Comprehensive investigation of SARS-CoV-2 intestinal pathogenesis in Drosophila.bioRxiv [Preprint]. 2025 Jun 25:2025.06.25.661044. doi: 10.1101/2025.06.25.661044. bioRxiv. 2025. PMID: 40667169 Free PMC article. Preprint.
-
Integrated analysis of RNA-seq datasets reveals novel targets and regulators of COVID-19 severity.Life Sci Alliance. 2024 Jan 23;7(4):e202302358. doi: 10.26508/lsa.202302358. Print 2024 Apr. Life Sci Alliance. 2024. PMID: 38262689 Free PMC article.
-
What do we know about the function of SARS-CoV-2 proteins?Front Immunol. 2023 Sep 18;14:1249607. doi: 10.3389/fimmu.2023.1249607. eCollection 2023. Front Immunol. 2023. PMID: 37790934 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

