Isolation and characterization of a Vibrio owensii phage phi50-12
- PMID: 36180722
- PMCID: PMC9525291
- DOI: 10.1038/s41598-022-20831-2
Isolation and characterization of a Vibrio owensii phage phi50-12
Abstract
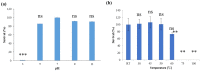
Vibrio owensii is a widely distributed marine vibrio species that causes acute hepatopancreatic necrosis in the larvae of Panulirus ornatus and Penaeus vannamei, and is also associated with Montipora white syndrome in corals. We characterized V. owensii GRA50-12 as a potent pathogen using phenotypic, biochemical, and zebrafish models. A virulent phage, vB_VowP_phi50-12 (phi50-12), belonging to the N4-like Podoviridae, was isolated from the same habitat as that of V. owensii GRA50-12 and characterized. This phage possesses a unique sequence with no similar hits in the public databases and has a short latent time (30 min), a large burst size (106 PFU/infected cell), and a wide range of pH and temperature stabilities. Moreover, phi50-12 also demonstrated a strong lysis ability against V. owensii GRA50-12. SDS-PAGE revealed at least nine structural proteins, four of which were confirmed using LC-MS/MS analysis. The size of the phi50-12 genome was 68,059 bp, with 38.5% G + C content. A total of 101 ORFs were annotated, with 17 ORFs having closely related counterparts in the N4-like vibrio phage. Genomic sequencing confirmed the absence of antibiotic resistance genes or virulence factors. Comparative studies have shown that phi50-12 has a unique genomic arrangement, except for the well-conserved core regions of the N4-like phages. Phylogenetic analysis demonstrated that it belonged to a group of smaller genomes of N4-like vibrio phages. The therapeutic effect in the zebrafish model suggests that phi50-12 could be a potential candidate for application in the treatment of V. owensii infection or as a biocontrol agent. However, further research must be carried out to confirm the efficacy of phage50-12.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

