Serine metabolism orchestrates macrophage polarization by regulating the IGF1-p38 axis
- PMID: 36180780
- PMCID: PMC9622887
- DOI: 10.1038/s41423-022-00925-7
Serine metabolism orchestrates macrophage polarization by regulating the IGF1-p38 axis
Abstract
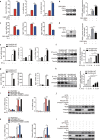
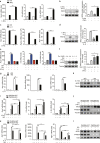
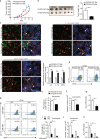
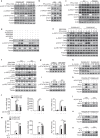
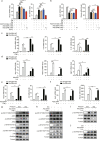
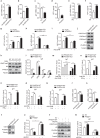
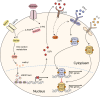
Serine metabolism is reportedly involved in immune cell functions, but whether and how serine metabolism regulates macrophage polarization remain largely unknown. Here, we show that suppressing serine metabolism, either by inhibiting the activity of the key enzyme phosphoglycerate dehydrogenase in the serine biosynthesis pathway or by exogenous serine and glycine restriction, robustly enhances the polarization of interferon-γ-activated macrophages (M(IFN-γ)) but suppresses that of interleukin-4-activated macrophages (M(IL-4)) both in vitro and in vivo. Mechanistically, serine metabolism deficiency increases the expression of IGF1 by reducing the promoter abundance of S-adenosyl methionine-dependent histone H3 lysine 27 trimethylation. IGF1 then activates the p38-dependent JAK-STAT1 axis to promote M(IFN-γ) polarization and suppress STAT6-mediated M(IL-4) activation. This study reveals a new mechanism by which serine metabolism orchestrates macrophage polarization and suggests the manipulation of serine metabolism as a therapeutic strategy for macrophage-mediated immune diseases.
Keywords: IGF1; Macrophage polarization; PHGDH; SAM; Serine metabolism; p38.
© 2022. The Author(s), under exclusive licence to CSI and USTC.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

