Plasma proteomic signature predicts who will get persistent symptoms following SARS-CoV-2 infection
- PMID: 36182629
- PMCID: PMC9515404
- DOI: 10.1016/j.ebiom.2022.104293
Plasma proteomic signature predicts who will get persistent symptoms following SARS-CoV-2 infection
Abstract
Background: The majority of those infected by ancestral Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) during the UK first wave (starting March 2020) did not require hospitalisation. Most had a short-lived mild or asymptomatic infection, while others had symptoms that persisted for weeks or months. We hypothesized that the plasma proteome at the time of first infection would reflect differences in the inflammatory response that linked to symptom severity and duration.
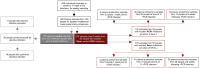
Methods: We performed a nested longitudinal case-control study and targeted analysis of the plasma proteome of 156 healthcare workers (HCW) with and without lab confirmed SARS-CoV-2 infection. Targeted proteomic multiple-reaction monitoring analysis of 91 pre-selected proteins was undertaken in uninfected healthcare workers at baseline, and in infected healthcare workers serially, from 1 week prior to 6 weeks after their first confirmed SARS-CoV-2 infection. Symptom severity and antibody responses were also tracked. Questionnaires at 6 and 12 months collected data on persistent symptoms.
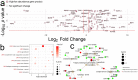
Findings: Within this cohort (median age 39 years, interquartile range 30-47 years), 54 healthcare workers (44% male) had PCR or antibody confirmed infection, with the remaining 102 (38% male) serving as uninfected controls. Following the first confirmed SARS-CoV-2 infection, perturbation of the plasma proteome persisted for up to 6 weeks, tracking symptom severity and antibody responses. Differentially abundant proteins were mostly coordinated around lipid, atherosclerosis and cholesterol metabolism pathways, complement and coagulation cascades, autophagy, and lysosomal function. The proteomic profile at the time of seroconversion associated with persistent symptoms out to 12 months. Data are available via ProteomeXchange with identifier PXD036590.
Interpretation: Our findings show that non-severe SARS-CoV-2 infection perturbs the plasma proteome for at least 6 weeks. The plasma proteomic signature at the time of seroconversion has the potential to identify which individuals are more likely to suffer from persistent symptoms related to SARS-CoV-2 infection.
Funding information: The COVIDsortium is supported by funding donated by individuals, charitable Trusts, and corporations including Goldman Sachs, Citadel and Citadel Securities, The Guy Foundation, GW Pharmaceuticals, Kusuma Trust, and Jagclif Charitable Trust, and enabled by Barts Charity with support from University College London Hospitals (UCLH) Charity. This work was additionally supported by the Translational Mass Spectrometry Research Group and the Biomedical Research Center (BRC) at Great Ormond Street Hospital.
Keywords: COVID-19; Post-acute sequelae of SARS-CoV-2 (PASC); Proteomics.
Copyright © 2022. Published by Elsevier B.V.
Conflict of interest statement
Declaration of interests RJB and DMA are members of the Global T cell Expert Consortium and have consulted for Oxford Immunotec outside the submitted work.
Figures




References
-
- World Health Organization. A clinical case definition of post COVID-19 condition by a Delphi consensus, 6 October 2021. 2021.
-
- Briggs A, Vassall A. Count the cost of disability caused by COVID-19. Nature. 2021;593:502–505. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

