Huc-MSC-derived exosomes modified with the targeting peptide of aHSCs for liver fibrosis therapy
- PMID: 36183106
- PMCID: PMC9526331
- DOI: 10.1186/s12951-022-01636-x
Huc-MSC-derived exosomes modified with the targeting peptide of aHSCs for liver fibrosis therapy
Abstract
Background: Effective therapeutics to stop or reverse liver fibrosis have not emerged, because these potential agents cannot specifically target activated hepatic stellate cells (aHSCs) or are frequently toxic to parenchymal cells. Human umbilical cord mesenchymal stem cell (Huc-MSC)-derived exosomes show promise in nanomedicine for the treatment of liver fibrosis. However, systemic injection showed that unmodified exosomes were mainly taken up by the mononuclear phagocyte system. The discovery of ligands that selectively bind to a specific target plays a crucial role in clinically relevant diagnostics and therapeutics. Herein, we aimed to identify the targeting peptide of aHSCs by screening a phage-displayed peptide library, and modify Huc-MSC-derived exosomes with the targeting peptide.
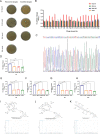
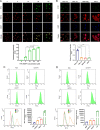
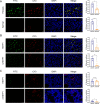
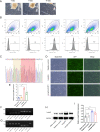
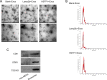
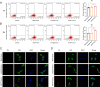
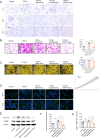
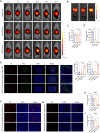
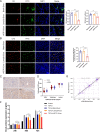
Results: In this study, we screened a phage-displayed peptide library by biopanning for peptides preferentially bound to HSC-T6 cells. The identified peptide, HSTP1, also exhibited better targeting ability to aHSCs in pathological sections of fibrotic liver tissues. Then, HSTP1 was fused with exosomal enriched membrane protein (Lamp2b) and was displayed on the surface of exosomes through genetic engineering technology. The engineered exosomes (HSTP1-Exos) could be more efficiently internalized by HSC-T6 cells and outperformed both unmodified exosomes (Blank-Exos) and Lamp2b protein overexpressed exosomes (Lamp2b + Exos) in enhancing the ability of exosomes to promote HSC-T6 reversion to a quiescent phenotype. In vivo results showed HSTP1-Exos could specifically target to the aHSC region after intravenous administration, as demonstrated by coimmunofluorescence with the typical aHSCs marker α-SMA, and enhance the therapeutic effect on liver fibrosis.
Conclusion: These results suggest that HSTP1 is a reliable targeting peptide that can specifically bind to aHSCs and that HSTP1-modified exosomes realize the precise treatment for aHSCs in complex liver tissue. We provide a novel strategy for clinical liver fibrosis therapy.
Keywords: Exosomes; Huc-MSCs; Liver fibrosis; Targeting peptide; aHSCs.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Du XS, Li HD, Yang XJ, Li JJ, Xu JJ, Chen Y, Xu QQ, Yang L, He CS, Huang C, et al. Wogonin attenuates liver fibrosis via regulating hepatic stellate cell activation and apoptosis. Int Immunopharmacol. 2019;75:105671. - PubMed
-
- Mejias M, Gallego J, Naranjo-Suarez S, Ramirez M, Pell N, Manzano A, Suñer C, Bartrons R, Mendez R, Fernandez M. Cpeb4 increases expression of PFKFB3 to induce glycolysis and activate mouse and human hepatic stellate cells, promoting liver fibrosis. Gastroenterology. 2020;159(1):273–288. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

