Regional epithelial cell diversity in the small intestine of pigs
- PMID: 36183288
- PMCID: PMC9831138
- DOI: 10.1093/jas/skac318
Regional epithelial cell diversity in the small intestine of pigs
Abstract
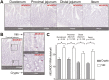
Understanding regional distribution and specialization of small intestinal epithelial cells is crucial for developing methods to control appetite, stress, and nutrient uptake in swine. To establish a better understanding of specific epithelial cells found across different regions of the small intestine in pigs, we utilized single-cell RNA sequencing (scRNA-seq) to recover and analyze epithelial cells from duodenum, jejunum, and ileum. Cells identified included crypt cells, enterocytes, BEST4 enterocytes, goblet cells, and enteroendocrine (EE) cells. EE cells were divided into two subsets based on the level of expression of the EE lineage commitment gene, NEUROD1. NEUROD1hi EE cells had minimal expression of hormone-encoding genes and were dissimilar to EE cells in humans and mice, indicating a subset of EE cells unique to pigs. Recently discovered BEST4 enterocytes were detected in both crypts and villi throughout the small intestine via in situ staining, unlike in humans, where BEST4 enterocytes are found only in small intestinal villi. Proximal-to-distal gradients of expression were noted for hormone-encoding genes in EE cells and nutrient transport genes in enterocytes via scRNA-seq, demonstrating regional specialization. Regional gene expression in EE cells and enterocytes was validated via quantitative PCR (qPCR) analysis of RNA isolated from epithelial cells of different small intestinal locations. Though many genes had similar patterns of regional expression when assessed by qPCR of total epithelial cells, some regional expression was only detected via scRNA-seq, highlighting advantages of scRNA-seq to deconvolute cell type-specific regional gene expression when compared to analysis of bulk samples. Overall, results provide new information on regional localization and transcriptional profiles of epithelial cells in the pig small intestine.
Keywords: BEST4 enterocyte; NEUROD1; enterochromaffin; enteroendocrine; porcine; single-cell RNA sequencing.
Plain language summary
Cells lining the intestinal tract (i.e., epithelial cells) provide a barrier to the outside environment but also play important specialized roles in nutrient absorption and secretion of mucus or hormones involved in controlling appetite and digestion. While similar cell types can be found throughout the small intestine, they have even more specialized function depending on region of the small intestine. Identification and characterization of intestinal epithelial cells are foundational to promoting pig intestinal health for optimal growth. Our research identified six types of epithelial cells across the small intestine of pigs. Enterocytes, an absorptive cell type, shared commonalities with human enterocytes, but a population of enteroendocrine cells, which secrete hormones, was unique to pigs. The location of certain epithelial cells in the intestine was identified and informed the relationship between various epithelial cell types. Overall, a clearer understanding of specific epithelial cells in the porcine intestine is provided, proving a critical foundation to further research aimed at maximizing pig intestinal health.
Published by Oxford University Press on behalf of the American Society of Animal Science 2022.
Conflict of interest statement
All opinions expressed in this article are of the authors' and do not necessarily reflect the policies and views of the supporting agencies.
Figures





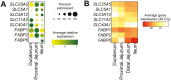
Similar articles
-
BEST4+ cells in the intestinal epithelium.Am J Physiol Cell Physiol. 2024 May 1;326(5):C1345-C1352. doi: 10.1152/ajpcell.00042.2024. Epub 2024 Apr 1. Am J Physiol Cell Physiol. 2024. PMID: 38557358 Free PMC article. Review.
-
Nutrient sensing by absorptive and secretory progenies of small intestinal stem cells.Am J Physiol Gastrointest Liver Physiol. 2017 Jun 1;312(6):G592-G605. doi: 10.1152/ajpgi.00416.2016. Epub 2017 Mar 23. Am J Physiol Gastrointest Liver Physiol. 2017. PMID: 28336548 Free PMC article.
-
Immunohistochemical characterization of the distribution of galectin-4 in porcine small intestine.J Histochem Cytochem. 2005 Feb;53(2):197-205. doi: 10.1369/jhc.4A6439.2005. J Histochem Cytochem. 2005. PMID: 15684332
-
Adult stem cells in the small intestine are intrinsically programmed with their location-specific function.Stem Cells. 2014 May;32(5):1083-91. doi: 10.1002/stem.1655. Stem Cells. 2014. PMID: 24496776
-
Glucose and amino acid in enterocyte: absorption, metabolism and maturation.Front Biosci (Landmark Ed). 2018 Mar 1;23(9):1721-1739. doi: 10.2741/4669. Front Biosci (Landmark Ed). 2018. PMID: 29293459 Review.
Cited by
-
From a hunger-regulating hormone to an antimicrobial peptide: gastrointestinal derived circulating endocrine hormone-peptide YY exerts exocrine antimicrobial effects against selective gut microbiota.Gut Microbes. 2024 Jan-Dec;16(1):2316927. doi: 10.1080/19490976.2024.2316927. Epub 2024 Feb 14. Gut Microbes. 2024. PMID: 38356283 Free PMC article. No abstract available.
-
Development of an intestinal epithelial cell line and organoids derived from the same swine and characterization of their antiviral responses.Biosci Microbiota Food Health. 2024;43(4):342-351. doi: 10.12938/bmfh.2024-0046. Epub 2024 May 28. Biosci Microbiota Food Health. 2024. PMID: 39364127 Free PMC article.
-
Development and characterization of segment-specific enteroids from the pig small intestine in Matrigel and transwell inserts: insights into susceptibility to porcine epidemic diarrhea Virus.Front Immunol. 2024 Sep 17;15:1451154. doi: 10.3389/fimmu.2024.1451154. eCollection 2024. Front Immunol. 2024. PMID: 39355235 Free PMC article.
-
Advances in single-cell transcriptomics in animal research.J Anim Sci Biotechnol. 2024 Aug 2;15(1):102. doi: 10.1186/s40104-024-01063-y. J Anim Sci Biotechnol. 2024. PMID: 39090689 Free PMC article. Review.
-
BEST4+ cells in the intestinal epithelium.Am J Physiol Cell Physiol. 2024 May 1;326(5):C1345-C1352. doi: 10.1152/ajpcell.00042.2024. Epub 2024 Apr 1. Am J Physiol Cell Physiol. 2024. PMID: 38557358 Free PMC article. Review.
References
-
- Borisova, M. A., Achasova K. M., Morozova K. N., Andreyeva E. N., Litvinova E. A., Ogienko A. A., Morozova M. V., Berkaeva M. B., Kiseleva E., and Kozhevnikova E. N... 2020. Mucin-2 knockout is a model of intercellular junction defects, mitochondrial damage and ATP depletion in the intestinal epithelium. Sci. Rep. 10:21135. doi:10.1038/s41598-020-78141-4 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 5030-32000-225-00D/United States Department of Agriculture-Agriculture Research Service
- Agricultural Research Service Research Participation Program
- Oak Ridge Institute for Science and Education
- United States Department of Energy and the United States Department of Agriculture
- DE-SC0014664/Oak Ridge Associated Universities
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

