Rapid genomic changes by mineralotropic hormones and kinase SIK inhibition drive coordinated renal Cyp27b1 and Cyp24a1 expression via CREB modules
- PMID: 36183832
- PMCID: PMC9637672
- DOI: 10.1016/j.jbc.2022.102559
Rapid genomic changes by mineralotropic hormones and kinase SIK inhibition drive coordinated renal Cyp27b1 and Cyp24a1 expression via CREB modules
Abstract
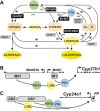
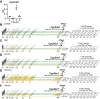
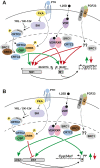
Vitamin D metabolism centers on kidney regulation of Cyp27b1 by mineralotropic hormones, including induction by parathyroid hormone (PTH), suppression by fibroblast growth factor 23 (FGF23) and 1,25-dihydroxyvitamin D3 (1,25(OH)2D3), and reciprocal regulations for Cyp24a1. This coordinated genomic regulation results in production of endocrine 1,25(OH)2D3, which, together with PTH and FGF23, controls mineral homeostasis. However, how these events are coordinated is unclear. Here, using in vivo chromatin immunoprecipitation sequencing in mouse kidney, we demonstrate that PTH activation rapidly induces increased recruitment of phosphorylated (p-133) CREB (pCREB) and its coactivators, CBP (CREB-binding protein) and CRTC2 (CREB-regulated transcription coactivator 2), to previously defined kidney-specific M1 and M21 enhancers near the Cyp27b1 gene. At distal enhancers of the Cyp24a1 gene, PTH suppression dismisses CBP with only minor changes in pCREB and CRTC2 occupancy, all of which correlate with decreased genomic activity and reduced transcripts. Treatment of mice with salt-inducible kinase inhibitors (YKL-05-099 and SK-124) yields rapid genomic recruitment of CRTC2 to Cyp27b1, limited interaction of CBP, and a transcriptional response for both Cyp27b1 and Cyp24a1 that mirrors the actions of PTH. Surprisingly, we find that 1,25(OH)2D3 suppression increases the occupancy of CRTC2 in the M1 enhancer, a novel observation for CRTC2 and 1,25(OH)2D3 action. Suppressive actions of 1,25(OH)2D3 and FGF23 at the Cyp27b1 gene are associated with reduced CBP recruitment at these CREB-module enhancers that disrupts full PTH induction. Our findings show that CRTC2 contributes to transcription of both Cyp27b1 and Cyp24a1, demonstrate salt-inducible kinase inhibition as a key modulator of vitamin D metabolism, and provide molecular insight into the coordinated mechanistic actions of PTH, FGF23, and 1,25(OH)2D3 in the kidney that regulate mineral homeostasis.
Keywords: 1,25(OH)(2)D(3); ChIP-Seq; Cyp24a1; Cyp27b1; FGF23; cytochrome P450; gene regulation; parathyroid hormone; salt-inducible kinases; vitamin D.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest M. N. W. has consulted for AstraZeneca, Guidepoint, and Galapagos and receives research support from Radius Health. All other authors declare no conflicts of interest with the contents of this article.
Figures








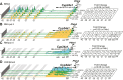

References
-
- Jones G., Strugnell S.A., DeLuca H.F. Current understanding of the molecular actions of vitamin D. Physiol. Rev. 1998;78:1193–1231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

