NSUN2-mediated mRNA m5C Modification Regulates the Progression of Hepatocellular Carcinoma
- PMID: 36183976
- PMCID: PMC10787115
- DOI: 10.1016/j.gpb.2022.09.007
NSUN2-mediated mRNA m5C Modification Regulates the Progression of Hepatocellular Carcinoma
Abstract
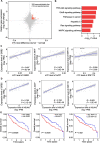
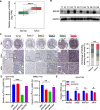
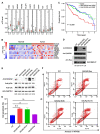
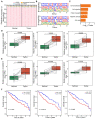
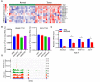
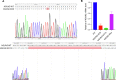
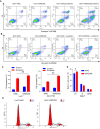
RNA modifications affect many biological processes and physiological diseases. The 5-methylcytosine (m5C) modification regulates the progression of multiple tumors. However, its characteristics and functions in hepatocellular carcinoma (HCC) remain largely unknown. Here, we found that HCC tissues had a higher m5C methylation level than the adjacent normal tissues. Transcriptome analysis revealed that the hypermethylated genes mainly participated in the phosphokinase signaling pathways, such as the Ras and PI3K-Akt pathways. The m5C methyltransferase NSUN2 was highly expressed in HCC tissues. Interestingly, the expression of many genes was positively correlated with the expression of NSUN2, including GRB2, RNF115, AATF, ADAM15, RTN3, and HDGF. Real-time PCR assays further revealed that the expression of the mRNAs of GRB2, RNF115, and AATF decreased significantly with the down-regulation of NSUN2 expression in HCC cells. Furthermore, NSUN2 could regulate the cellular sensitivity of HCC cells to sorafenib via modulating the Ras signaling pathway. Moreover, knocking down NSUN2 caused cell cycle arrest. Taken together, our study demonstrates the vital role of NSUN2 in the progression of HCC.
Keywords: 5-methylcytosine; Hepatocellular carcinoma; NSUN2; Ras pathway; Sorafenib.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
The RNA M5C methyltransferase NSUN2 promotes progression of hepatocellular carcinoma by enhancing PKM2-mediated glycolysis.Cell Death Dis. 2025 Feb 9;16(1):82. doi: 10.1038/s41419-025-07414-5. Cell Death Dis. 2025. PMID: 39924557 Free PMC article.
-
NSUN2 promotes lung adenocarcinoma progression through stabilizing PIK3R2 mRNA in an m5C-dependent manner.Mol Carcinog. 2024 May;63(5):962-976. doi: 10.1002/mc.23701. Epub 2024 Feb 27. Mol Carcinog. 2024. PMID: 38411298
-
NSUN2 regulates Wnt signaling pathway depending on the m5C RNA modification to promote the progression of hepatocellular carcinoma.Oncogene. 2024 Nov;43(47):3469-3482. doi: 10.1038/s41388-024-03184-0. Epub 2024 Oct 7. Oncogene. 2024. PMID: 39375506
-
NSUN2-mediated RNA 5-methylcytosine promotes esophageal squamous cell carcinoma progression via LIN28B-dependent GRB2 mRNA stabilization.Oncogene. 2021 Sep;40(39):5814-5828. doi: 10.1038/s41388-021-01978-0. Epub 2021 Aug 3. Oncogene. 2021. PMID: 34345012 Free PMC article.
-
Upregulation of lncRNA NIFK-AS1 in hepatocellular carcinoma by m6A methylation promotes disease progression and sorafenib resistance.Hum Cell. 2021 Nov;34(6):1800-1811. doi: 10.1007/s13577-021-00587-z. Epub 2021 Aug 10. Hum Cell. 2021. PMID: 34374933 Review.
Cited by
-
Underexplored reciprocity between genome-wide methylation status and long non-coding RNA expression reflected in breast cancer research: potential impacts for the disease management in the framework of 3P medicine.EPMA J. 2023 May 22;14(2):249-273. doi: 10.1007/s13167-023-00323-7. eCollection 2023 Jun. EPMA J. 2023. PMID: 37275549 Free PMC article. Review.
-
RNA m5C methylation: a potential modulator of innate immune pathways in hepatocellular carcinoma.Front Immunol. 2024 May 13;15:1362159. doi: 10.3389/fimmu.2024.1362159. eCollection 2024. Front Immunol. 2024. PMID: 38807595 Free PMC article. Review.
-
Clinical significance of RNA methylation in hepatocellular carcinoma.Cell Commun Signal. 2024 Apr 2;22(1):204. doi: 10.1186/s12964-024-01595-w. Cell Commun Signal. 2024. PMID: 38566136 Free PMC article. Review.
-
Role of m5C methylation in digestive system tumors (Review).Mol Med Rep. 2025 Jun;31(6):142. doi: 10.3892/mmr.2025.13507. Epub 2025 Apr 4. Mol Med Rep. 2025. PMID: 40183387 Free PMC article.
-
The multifaceted role of m5C RNA methylation in digestive system tumorigenesis.Front Cell Dev Biol. 2025 Mar 6;13:1533148. doi: 10.3389/fcell.2025.1533148. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40114967 Free PMC article. Review.
References
-
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. - PubMed
-
- Sia D., Villanueva A., Friedman S.L., Llovet J.M. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152:745–761. - PubMed
-
- Akula S.M., Abrams S.L., Steelman L.S., Emma M.R., Augello G., Cusimano A., et al. RAS/RAF/MEK/ERK, PI3K/PTEN/AKT/mTORC1 and TP53 pathways and regulatory miRs as therapeutic targets in hepatocellular carcinoma. Expert Opin Ther Targets. 2019;23:915–929. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

