Research progress of CRISPR-based biosensors and bioassays for molecular diagnosis
- PMID: 36185462
- PMCID: PMC9524266
- DOI: 10.3389/fbioe.2022.986233
Research progress of CRISPR-based biosensors and bioassays for molecular diagnosis
Abstract
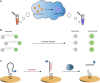
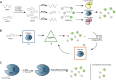
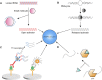
CRISPR/Cas technology originated from the immune mechanism of archaea and bacteria and was awarded the Nobel Prize in Chemistry in 2020 for its success in gene editing. Molecular diagnostics is highly valued globally for its development as a new generation of diagnostic technology. An increasing number of studies have shown that CRISPR/Cas technology can be integrated with biosensors and bioassays for molecular diagnostics. CRISPR-based detection has attracted much attention as highly specific and sensitive sensors with easily programmable and device-independent capabilities. The nucleic acid-based detection approach is one of the most sensitive and specific diagnostic methods. With further research, it holds promise for detecting other biomarkers such as small molecules and proteins. Therefore, it is worthwhile to explore the prospects of CRISPR technology in biosensing and summarize its application strategies in molecular diagnostics. This review provides a synopsis of CRISPR biosensing strategies and recent advances from nucleic acids to other non-nucleic small molecules or analytes such as proteins and presents the challenges and perspectives of CRISPR biosensors and bioassays.
Keywords: CRISPR/Cas; biosensor; molecular diagnostics; non-nucleic-acid analytes; nucleic acid detection.
Copyright © 2022 Chen, Shen, Wang, Gu, Zhao, Lin, Liu, Cai, Mushtaq, Jia, Wan and Yan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer HL declared a shared parent affiliation with the author YC at the time of review.
Figures

Similar articles
-
Advancing sensing technology with CRISPR: From the detection of nucleic acids to a broad range of analytes - A review.Anal Chim Acta. 2021 Nov 15;1185:338848. doi: 10.1016/j.aca.2021.338848. Epub 2021 Jul 13. Anal Chim Acta. 2021. PMID: 34711331 Review.
-
Recent Advances in CRISPR/Cas-Based Biosensors for Protein Detection.Bioengineering (Basel). 2022 Sep 28;9(10):512. doi: 10.3390/bioengineering9100512. Bioengineering (Basel). 2022. PMID: 36290480 Free PMC article. Review.
-
Novel non-nucleic acid targets detection strategies based on CRISPR/Cas toolboxes: A review.Biosens Bioelectron. 2022 Nov 1;215:114559. doi: 10.1016/j.bios.2022.114559. Epub 2022 Jul 13. Biosens Bioelectron. 2022. PMID: 35917610 Review.
-
Recent progress on the CRISPR/Cas system in optical biosensors.Anal Methods. 2024 Feb 8;16(6):798-816. doi: 10.1039/d3ay02147e. Anal Methods. 2024. PMID: 38259224 Review.
-
CRISPR-Based Biosensing Strategies: Technical Development and Application Prospects.Annu Rev Anal Chem (Palo Alto Calif). 2023 Jun 14;16(1):311-332. doi: 10.1146/annurev-anchem-090822-014725. Epub 2023 Apr 5. Annu Rev Anal Chem (Palo Alto Calif). 2023. PMID: 37018798 Review.
Cited by
-
Advances in cell-based biosensors: Transforming food flavor evaluation with novel approaches.Food Chem X. 2025 Mar 1;26:102336. doi: 10.1016/j.fochx.2025.102336. eCollection 2025 Feb. Food Chem X. 2025. PMID: 40115496 Free PMC article. Review.
-
Molecular Diagnosis and Cancer Prognosis-A Concise Review.Diagnostics (Basel). 2023 Feb 17;13(4):766. doi: 10.3390/diagnostics13040766. Diagnostics (Basel). 2023. PMID: 36832253 Free PMC article. Review.
-
LAMP-CRISPR/Cas12a-based impedimetric biosensor powered by Fe3O4@Au-(S-polyA-S)-Au for detection of SARS-CoV-2.Mikrochim Acta. 2024 Oct 3;191(11):644. doi: 10.1007/s00604-024-06688-4. Mikrochim Acta. 2024. PMID: 39361061
-
Rapid detection of Lys-gingipain using fluorogenic peptide substrate for diagnosis of periodontitis.J Dent Sci. 2025 Apr;20(2):802-810. doi: 10.1016/j.jds.2024.09.025. Epub 2024 Oct 15. J Dent Sci. 2025. PMID: 40224085 Free PMC article.
-
CRISPR Diagnostics for Quantification and Rapid Diagnosis of Myotonic Dystrophy Type 1 Repeat Expansion Disorders.ACS Synth Biol. 2024 Dec 20;13(12):3926-3935. doi: 10.1021/acssynbio.4c00265. Epub 2024 Nov 20. ACS Synth Biol. 2024. PMID: 39565688 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources

