Single-cell transcriptome profiling of the human endometrium of patients with recurrent implantation failure
- PMID: 36185612
- PMCID: PMC9516226
- DOI: 10.7150/thno.74053
Single-cell transcriptome profiling of the human endometrium of patients with recurrent implantation failure
Abstract
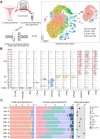
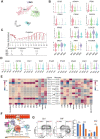
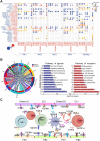
Introduction: Despite great advances in assisted reproductive technology (ART), recurrent implantation failure (RIF) cannot be effectively avoided. Notably, cellular characteristics and communication that regulate endometrial receptivity and differentiation, and its disorders in RIF at window of implantation (WOI) remain rudimentary. Objectives: In this study, we profiled the endometrial cells present at the WOI timing in RIF patients and healthy controls using single-cell RNA sequencing (scRNA-seq) and provided a detailed molecular and cellular map of a healthy and RIF endometrium at the WOI. Method: In the current study, the endometrium from RIF patient (n = 6; age range, 32 - 35 years) and control (Ctrl) (n = 3; age range, 29 - 35 years) groups were studied at a single-cell resolution. single-cell RNA-seq and analysis were performed on the endometrium of patients with RIF and Ctrl. Immunofluorescence, flow cytometry assays, and quantitative real-time polymerase chain reaction (qRT-PCR) were performed to verify cellular identity and function. Results: We profiled the transcriptomes of 60222 primary human endometrial cells isolated from control and RIF patients at a single-cell resolution. We discovered dramatic differential expression of endometrial receptivity-related genes in four major endometrial fibroblast-like cells from RIF patients compared to the control endometrium. We observed that CD49a+CXCR4+NK cells were diminished in proportion with RIF. The decrease in subset of CD63highPGRhigh endometrial epithelial cells with high levels of progesterone receptor, autophagy and exosomes should contribute to the decrease in subset of NK cells. Additionally, we characterized aberrant molecular and cellular characteristics and endometrial cell-cell communication disorders in RIF patients. Conclusion: Our study provides deeper insights into endometrial microenvironment disorder of RIF that are potentially applicable to improving the etiological diagnosis and therapeutics of unexplained RIF.
Keywords: NK cell; endometrial fibroblast-like cell; endometrial receptivity; epithelial cell; recurrent implantation failure.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures





Similar articles
-
Varied cellular abnormalities in thin vs. normal endometrium in recurrent implantation failure by single-cell transcriptomics.Reprod Biol Endocrinol. 2024 Jul 31;22(1):90. doi: 10.1186/s12958-024-01263-1. Reprod Biol Endocrinol. 2024. PMID: 39085925 Free PMC article.
-
Endometrial transcriptome profiling of patients with recurrent implantation failure during hormone replacement therapy cycles.Front Endocrinol (Lausanne). 2024 Jan 30;14:1292723. doi: 10.3389/fendo.2023.1292723. eCollection 2023. Front Endocrinol (Lausanne). 2024. PMID: 38352249 Free PMC article.
-
Friend leukemia integration 1 overexpression decreases endometrial receptivity and induces embryo implantation failure by promoting PART1 transcription in the endometrial epithelial cells.PeerJ. 2023 Sep 26;11:e16105. doi: 10.7717/peerj.16105. eCollection 2023. PeerJ. 2023. PMID: 37780395 Free PMC article.
-
The Contribution of Proteomics in Understanding Endometrial Protein Expression in Women with Recurrent Implantation Failure.J Clin Med. 2024 Apr 8;13(7):2145. doi: 10.3390/jcm13072145. J Clin Med. 2024. PMID: 38610911 Free PMC article. Review.
-
Human Endometrial Transcriptomics: Implications for Embryonic Implantation.Cold Spring Harb Perspect Med. 2015 Mar 27;5(7):a022996. doi: 10.1101/cshperspect.a022996. Cold Spring Harb Perspect Med. 2015. PMID: 25818663 Free PMC article. Review.
Cited by
-
Autophagy Inhibition in Trophoblasts Induces Aberrant Shift in CXCR4+ Decidual NK Cell Phenotype Leading to Pregnancy Loss.J Clin Med. 2023 Dec 4;12(23):7491. doi: 10.3390/jcm12237491. J Clin Med. 2023. PMID: 38068544 Free PMC article.
-
Endometrial proteomic profile of patients with repeated implantation failure.Front Endocrinol (Lausanne). 2023 Jul 31;14:1144393. doi: 10.3389/fendo.2023.1144393. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37583433 Free PMC article.
-
Construction of hub transcription factor-microRNAs-messenger RNA regulatory network in recurrent implantation failure.J Assist Reprod Genet. 2024 Jan;41(1):3-13. doi: 10.1007/s10815-023-02947-0. Epub 2023 Oct 25. J Assist Reprod Genet. 2024. PMID: 37878219 Free PMC article.
-
Enhancing endometrial receptivity: the roles of human chorionic gonadotropin in autophagy and apoptosis regulation in endometrial stromal cells.Reprod Biol Endocrinol. 2024 Apr 4;22(1):37. doi: 10.1186/s12958-024-01205-x. Reprod Biol Endocrinol. 2024. PMID: 38576003 Free PMC article.
-
An integrated single-cell reference atlas of the human endometrium.Nat Genet. 2024 Sep;56(9):1925-1937. doi: 10.1038/s41588-024-01873-w. Epub 2024 Aug 28. Nat Genet. 2024. PMID: 39198675 Free PMC article.
References
-
- Diedrich K, Fauser BC, Devroey P, Griesinger G, Evian Annual Reproduction Workshop G. The role of the endometrium and embryo in human implantation. Hum Reprod Update. 2007;13:365–77. - PubMed
-
- Coughlan C, Ledger W, Wang Q, Liu F, Demirol A, Gurgan T. et al. Recurrent implantation failure: definition and management. Reprod Biomed Online. 2014;28:14–38. - PubMed
-
- Wang W, Vilella F, Alama P, Moreno I, Mignardi M, Isakova A. et al. Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat Med. 2020;26:1644–53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

