Fungal and bacterial oxylipins are signals for intra- and inter-cellular communication within plant disease
- PMID: 36186042
- PMCID: PMC9524268
- DOI: 10.3389/fpls.2022.823233
Fungal and bacterial oxylipins are signals for intra- and inter-cellular communication within plant disease
Abstract
Lipids are central at various stages of host-pathogen interactions in determining virulence and modulating plant defense. Free fatty acids may act as substrates for oxidizing enzymes [e.g., lipoxygenases (LOXs) and dioxygenases (DOXs)] that synthesize oxylipins. Fatty acids and oxylipins function as modulators of several pathways in cell-to-cell communication; their structural similarity among plant, fungal, and bacterial taxa suggests potential in cross-kingdom communication. We provide a prospect of the known role of fatty acids and oxylipins in fungi and bacteria during plant-pathogen interactions. In the pathogens, oxylipin-mediated signaling pathways are crucial both in development and host infection. Here, we report on case studies suggesting that oxylipins derived from oleic, linoleic, and linolenic acids are crucial in modulating the pathogenic lifestyle in the host plant. Intriguingly, overlapping (fungi-plant/bacteria-plant) results suggest that different inter-kingdom pathosystems use similar lipid signals to reshape the lifestyle of the contenders and occasionally determine the outcome of the challenge.
Keywords: Aspergillus spp; Fusarium spp; Olea europaea L.; Xylella fastidiosa; Zea mays (L); lipids; oxylipins.
Copyright © 2022 Beccaccioli, Pucci, Salustri, Scortichini, Zaccaria, Momeni, Loreti, Reverberi and Scala.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
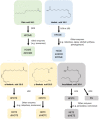
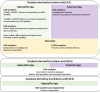
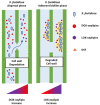
References
-
- Ambaw Y. A., Pagac M. P., Irudayaswamy A. S., Raida M., Bendt A. K., Torta F. T., et al. . (2021). Host/malassezia interaction: a quantitative, non-invasive method profiling oxylipin production associates human skin eicosanoids with Malassezia. Metabolites 11, 700. 10.3390/metabo11100700 - DOI - PMC - PubMed
-
- Battilani P., Lanubile A., Scala V., Reverberi M., Gregori R., Falavigna C., et al. . (2018). Oxylipins from both pathogen and host antagonize jasmonic acid-mediated defence via the 9-lipoxygenase pathway in Fusarium verticillioides infection of maize. Mol. Plant Pathol. 19, 2162–2176. 10.1111/mpp.12690 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

