Screening and validation of platelet activation-related lncRNAs as potential biomarkers for prognosis and immunotherapy in gastric cancer patients
- PMID: 36186426
- PMCID: PMC9515443
- DOI: 10.3389/fgene.2022.965033
Screening and validation of platelet activation-related lncRNAs as potential biomarkers for prognosis and immunotherapy in gastric cancer patients
Abstract
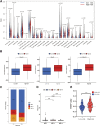
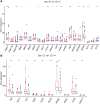
Background: Platelets (PLT) have a significant effect in promoting cancer progression and hematogenous metastasis. However, the effect of platelet activation-related lncRNAs (PLT-related lncRNAs) in gastric cancer (GC) is still poorly understood. In this study, we screened and validated PLT-related lncRNAs as potential biomarkers for prognosis and immunotherapy in GC patients. Methods: We obtained relevant datasets from the Cancer Genome Atlas (TCGA) and Gene Ontology (GO) Resource Database. Pearson correlation analysis was used to identify PLT-related lncRNAs. By using the univariate, least absolute shrinkage and selection operator (LASSO) Cox regression analyses, we constructed the PLT-related lncRNAs model. Kaplan-Meier survival analysis, univariate, multivariate Cox regression analysis, and nomogram were used to verify the model. The Gene Set Enrichment Analysis (GSEA), drug screening, tumor immune microenvironment analysis, epithelial-mesenchymal transition (EMT), and DNA methylation regulators correlation analysis were performed in the high- and low-risk groups. Patients were regrouped based on the risk model, and candidate compounds and immunotherapeutic responses aimed at GC subgroups were also identified. The expression of seven PLT-related lncRNAs was validated in clinical medical samples using quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Results: In this study, a risk prediction model was established using seven PLT-related lncRNAs -(AL355574.1, LINC01697, AC002401.4, AC129507.1, AL513123.1, LINC01094, and AL356417.2), whose expression were validated in GC patients. Kaplan-Meier survival analysis, the receiver operating characteristic (ROC) curve analysis, univariate, multivariate Cox regression analysis verified the accuracy of the model. We screened multiple targeted drugs for the high-risk patients. Patients in the high-risk group had a poorer prognosis since low infiltration of immune killer cells, activation of immunosuppressive pathways, and poor response to immunotherapy. In addition, we revealed a close relationship between risk scores and EMT and DNA methylation regulators. The nomogram based on risk score suggested a good ability to predict prognosis and high clinical benefits. Conclusion: Our findings provide new insights into how PLT-related lncRNAs biomarkers affect prognosis and immunotherapy. Also, these lncRNAs may become potential biomarkers and therapeutic targets for GC patients.
Keywords: gastric cancer; immunotherapy; lncRNA; platelet; prognosis.
Copyright © 2022 Yuan, Jia, Xing, Wang, Liu, Liu and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
LinkOut - more resources
Full Text Sources
Miscellaneous

