From classical mendelian randomization to causal networks for systematic integration of multi-omics
- PMID: 36186433
- PMCID: PMC9520987
- DOI: 10.3389/fgene.2022.990486
From classical mendelian randomization to causal networks for systematic integration of multi-omics
Abstract
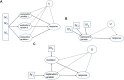
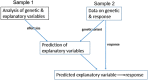
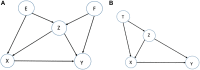
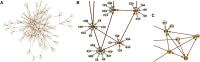
The number of studies with information at multiple biological levels of granularity, such as genomics, proteomics, and metabolomics, is increasing each year, and a biomedical questaion is how to systematically integrate these data to discover new biological mechanisms that have the potential to elucidate the processes of health and disease. Causal frameworks, such as Mendelian randomization (MR), provide a foundation to begin integrating data for new biological discoveries. Despite the growing number of MR applications in a wide variety of biomedical studies, there are few approaches for the systematic analysis of omic data. The large number and diverse types of molecular components involved in complex diseases interact through complex networks, and classical MR approaches targeting individual components do not consider the underlying relationships. In contrast, causal network models established in the principles of MR offer significant improvements to the classical MR framework for understanding omic data. Integration of these mostly distinct branches of statistics is a recent development, and we here review the current progress. To set the stage for causal network models, we review some recent progress in the classical MR framework. We then explain how to transition from the classical MR framework to causal networks. We discuss the identification of causal networks and evaluate the underlying assumptions. We also introduce some tests for sensitivity analysis and stability assessment of causal networks. We then review practical details to perform real data analysis and identify causal networks and highlight some of the utility of causal networks. The utilities with validated novel findings reveal the full potential of causal networks as a systems approach that will become necessary to integrate large-scale omic data.
Keywords: causal networks; classical MR; multiomic integration; principles of mendelian randomization; stability of causal networks; systems approach; systems biology.
Copyright © 2022 Yazdani, Yazdani, Mendez-Giraldez, Samiei, Kosorok and Schaid.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Abdi H., Valentin D. (2007). Multiple correspondence analysis. Encycl. Meas. Stat. 2, 65–66.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

