Sensitive fluorescence biosensor for SARS-CoV-2 nucleocapsid protein detection in cold-chain food products based on DNA circuit and g-CNQDs@Zn-MOF
- PMID: 36186577
- PMCID: PMC9510831
- DOI: 10.1016/j.lwt.2022.114032
Sensitive fluorescence biosensor for SARS-CoV-2 nucleocapsid protein detection in cold-chain food products based on DNA circuit and g-CNQDs@Zn-MOF
Abstract
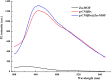
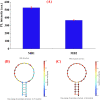
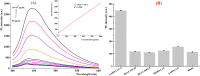
SARS-CoV-2 isolation from cold-chain food products confirms the possibility of outbreaks through cold-chain food products. RNA extraction combined with RT-PCR is the primary method currently utilized for the detection of SARS-CoV-2. However, the requirement of hours of analytical time and the high price of RT-PCR hinder its worldwide implementation in food supervision. Here, we report a fluorescence biosensor for detection of SARS-CoV-2 N protein. The fluorescence biosensor was fabricated by aptamer-based conformational entropy-driven circuit where molecular beacon strands were labeled with graphitic carbon nitrides quantum dots@Zn-metal-organic framework (g-CNQDs@Zn-MOF) and Dabcyl. The detection of the N protein was achieved via swabbing followed by competitive assay using a fixed amount of N-48 aptamers in the analytical system. A fluorescence emission spectrum was employed for the detection. The detection limit of our fluorescence biosensor was 1.0 pg/mL for SARS-CoV-2 N protein, indicating very excellent sensitivity. The fluorescence biosensor did not exhibit significant cross-reactivity with other N proteins. Finally, the biosensor was successfully applied for the detection of SARS-CoV-2 N protein in actual cold-chain food products showing same excellent accuracy as RT-PCR method. Thus, our fluorescence biosensor is a promising analytical tool for rapid and sensitive detection of SARS-CoV-2 N protein.
Keywords: Aptamer; Cold-chain food product; Conformational entropy-driven circuit; G-CNQDs@Zn-MOF; SARS-CoV-2.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
A ratiometric fluorescent sensor based on g-CNQDs@Zn-MOF for the sensitive detection of riboflavin via FRET.Spectrochim Acta A Mol Biomol Spectrosc. 2021 Feb 5;246:119004. doi: 10.1016/j.saa.2020.119004. Epub 2020 Oct 6. Spectrochim Acta A Mol Biomol Spectrosc. 2021. PMID: 33070014
-
Voltammetric-based immunosensor for the detection of SARS-CoV-2 nucleocapsid antigen.Mikrochim Acta. 2021 May 26;188(6):199. doi: 10.1007/s00604-021-04867-1. Mikrochim Acta. 2021. PMID: 34041585 Free PMC article.
-
Quantitative and sensitive detection of SARS coronavirus nucleocapsid protein using quantum dots-conjugated RNA aptamer on chip.J Chem Technol Biotechnol. 2011 Dec;86(12):1475-1479. doi: 10.1002/jctb.2721. Epub 2011 Oct 3. J Chem Technol Biotechnol. 2011. PMID: 32336860 Free PMC article.
-
SARS-CoV-2 virus label-free electrochemical nanohybrid MIP-aptasensor based on Ni3(BTC)2 MOF as a high-performance surface substrate.Mikrochim Acta. 2022 Jul 19;189(8):287. doi: 10.1007/s00604-022-05357-8. Mikrochim Acta. 2022. PMID: 35852630 Free PMC article.
-
Rapid field determination of SARS-CoV-2 by a colorimetric and fluorescent dual-functional lateral flow immunoassay biosensor.Sens Actuators B Chem. 2022 Jan 15;351:130897. doi: 10.1016/j.snb.2021.130897. Epub 2021 Oct 9. Sens Actuators B Chem. 2022. PMID: 34658530 Free PMC article.
Cited by
-
Sensing Characteristics of SARS-CoV-2 Spike Protein Using Aptamer-Functionalized Si-Based Electrolyte-Gated Field-Effect Transistor (EGT).Biosensors (Basel). 2024 Feb 26;14(3):124. doi: 10.3390/bios14030124. Biosensors (Basel). 2024. PMID: 38534231 Free PMC article.
-
Development and Application of an SPR Nanobiosensor Based on AuNPs for the Detection of SARS-CoV-2 on Food Surfaces.Biosensors (Basel). 2022 Dec 1;12(12):1101. doi: 10.3390/bios12121101. Biosensors (Basel). 2022. PMID: 36551068 Free PMC article.
-
Hierarchical Nanobiosensors at the End of the SARS-CoV-2 Pandemic.Biosensors (Basel). 2024 Feb 18;14(2):108. doi: 10.3390/bios14020108. Biosensors (Basel). 2024. PMID: 38392027 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
