SARS-CoV-2 detection methods: A comprehensive review
- PMID: 36186678
- PMCID: PMC9512523
- DOI: 10.1016/j.sjbs.2022.103465
SARS-CoV-2 detection methods: A comprehensive review
Abstract
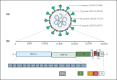
The ongoing novel COVID-19 has remained the center of attention, since its declaration as a pandemic in March 2020, due to its rapid and uncontrollable worldwide spread. Diagnostic tests are the first line of defense against the transmission of this infectious disease among individuals, with reverse-transcription quantitative polymerase chain reaction (RT-qPCR) being the approved gold standard for showing high sensitivity and specificity in detecting SARS-CoV-2. However, alternative tests are being invested due to the global demand for facilities, reagents, and healthcare workers needed for rapid population-based testing. Also, the rapid evolution of the viral genome and the emergence of new variants necessitates updating the existing methods. Scientists are aiming to improve tests to be affordable, simple, fast, and at the same time accurate, and efficient, as well as friendly user testing. The current diagnostic methods are either molecular-based that detect nucleic acids abundance, like RT-qPCR and reverse-transcription loop-mediated isothermal amplification (RT-LAMP); or immunologically based that detect the presence of antigens or antibodies in patients' specimens, like enzyme-linked immunosorbent assay (ELISA), lateral flow assay (LFA), chemiluminescent immunoassay (CLIA), and neutralization assay. In addition to these strategies, sensor-based or CRISPR applications are promising tools for the rapid detection of SARS-CoV-2. This review summarizes the most recent updates on the SARS-CoV-2 detection methods with their limitations. It will guide researchers, epidemiologists, and clinicians in identifying a more rapid, reliable, and sensitive method of diagnosing SARS-CoV-2 including the most recent variant of concern Omicron.
Keywords: COVID-19; Detection methods; RT-qPCR; SARS-CoV-2; Serology; Specimens; Variants.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Abdullah M., Sudrajat D.G., Muzellina V.N., Kurniawan J., Rizka A., Utari A.P., Pribadi R.R., Idrus M.F., Yusra Y., Meilany S., Surandy A., Shatri H., Rinaldi I., Pitoyo C.W., Renaldi K. The value of anal swab RT-PCR for COVID-19 diagnosis in adult Indonesian patients. BMJ Open Gastroenterol. 2021;8 - PMC - PubMed
-
- Abe, K. T., Li, Z., Samson, R., Samavarchi-Tehrani, P., Valcourt, E.J., Wood, H., Budylowski, P., Dupuis, A.P., 2ND, Girardin, R.C., Rathod, B., Wang, J.H., Barrios-Rodiles, M., Colwill, K., Mcgeer, A.J., Mubareka, S., Gommerman, J.L., Durocher, Y., Ostrowski, M., Mcdonough, K.A., Drebot, M.A., Drews, S.J., Rini, J.M., Gingras, A.C., 2020. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight, 5. - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

