Comparison of epidemiological characteristics and transmissibility of different strains of COVID-19 based on the incidence data of all local outbreaks in China as of March 1, 2022
- PMID: 36187650
- PMCID: PMC9521362
- DOI: 10.3389/fpubh.2022.949594
Comparison of epidemiological characteristics and transmissibility of different strains of COVID-19 based on the incidence data of all local outbreaks in China as of March 1, 2022
Abstract
Background: The epidemiological characteristics and transmissibility of Coronavirus Disease 2019 (COVID-19) may undergo changes due to the mutation of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) strains. The purpose of this study is to compare the differences in the outbreaks of the different strains with regards to aspects such as epidemiological characteristics, transmissibility, and difficulties in prevention and control.
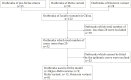
Methods: COVID-19 data from outbreaks of pre-Delta strains, the Delta variant and Omicron variant, were obtained from the Chinese Center for Disease Control and Prevention (CDC). Case data were collected from China's direct-reporting system, and the data concerning outbreaks were collected by on-site epidemiological investigators and collated by the authors of this paper. Indicators such as the effective reproduction number (R eff), time-dependent reproduction number (R t), rate of decrease in transmissibility (RDT), and duration from the illness onset date to the diagnosed date (D ID )/reported date (D IR ) were used to compare differences in transmissibility between pre-Delta strains, Delta variants and Omicron variants. Non-parametric tests (namely the Kruskal-Wallis H and Mean-Whitney U tests) were used to compare differences in epidemiological characteristics and transmissibility between outbreaks of different strains. P < 0.05 indicated that the difference was statistically significant.
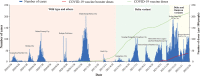
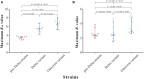
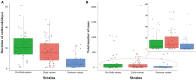
Results: Mainland China has maintained a "dynamic zero-out strategy" since the first case was reported, and clusters of outbreaks have occurred intermittently. The strains causing outbreaks in mainland China have gone through three stages: the outbreak of pre-Delta strains, the outbreak of the Delta variant, and outbreaks involving the superposition of Delta and Omicron variant strains. Each outbreak of pre-Delta strains went through two stages: a rising stage and a falling stage, Each outbreak of the Delta variant and Omicron variant went through three stages: a rising stage, a platform stage and a falling stage. The maximum R eff value of Omicron variant outbreaks was highest (median: 6.7; ranged from 5.3 to 8.0) and the differences were statistically significant. The RDT value of outbreaks involving pre-Delta strains was smallest (median: 91.4%; [IQR]: 87.30-94.27%), and the differences were statistically significant. The D ID and D IR for all strains was mostly in a range of 0-2 days, with more than 75%. The range of duration for outbreaks of pre-Delta strains was the largest (median: 20 days, ranging from 1 to 61 days), and the differences were statistically significant.
Conclusion: With the evolution of the virus, the transmissibility of the variants has increased. The transmissibility of the Omicron variant is higher than that of both the pre-Delta strains and the Delta variant, and is more difficult to suppress. These findings provide us with get a more clear and precise picture of the transmissibility of the different variants in the real world, in accordance with the findings of previous studies. R eff is more suitable than R t for assessing the transmissibility of the disease during an epidemic outbreak.
Keywords: Delta variant; Omicron variant; SARS-CoV-2; effective reproduction number; transmissibility; variant.
Copyright © 2022 Niu, Luo, Yang, Abudurusuli, Wang, Zhao, Rui, Li, Deng, Liu, Zhang, Li, Liu, Li, Huang, Yang, Wang, Chen and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
A comparative study on epidemiological characteristics, transmissibility, and pathogenicity of three COVID-19 outbreaks caused by different variants.Int J Infect Dis. 2023 Sep;134:78-87. doi: 10.1016/j.ijid.2023.01.039. Epub 2023 Feb 1. Int J Infect Dis. 2023. PMID: 36736993 Free PMC article.
-
Epidemiological characteristics and transmission dynamics of the outbreak caused by the SARS-CoV-2 Omicron variant in Shanghai, China: a descriptive study.medRxiv [Preprint]. 2022 Jun 18:2022.06.11.22276273. doi: 10.1101/2022.06.11.22276273. medRxiv. 2022. Update in: Lancet Reg Health West Pac. 2022 Dec;29:100592. doi: 10.1016/j.lanwpc.2022.100592. PMID: 35765564 Free PMC article. Updated. Preprint.
-
The rapid and efficient strategy for SARS-CoV-2 Omicron transmission control: analysis of outbreaks at the city level.Infect Dis Poverty. 2022 Nov 24;11(1):114. doi: 10.1186/s40249-022-01043-2. Infect Dis Poverty. 2022. PMID: 36434701 Free PMC article.
-
Is the SARS CoV-2 Omicron Variant Deadlier and More Transmissible Than Delta Variant?Int J Environ Res Public Health. 2022 Apr 11;19(8):4586. doi: 10.3390/ijerph19084586. Int J Environ Res Public Health. 2022. PMID: 35457468 Free PMC article. Review.
-
Comparison of Omicron and Delta Variants of SARS-CoV-2: A Systematic Review of Current Evidence.Infect Disord Drug Targets. 2024;24(7):e050324227686. doi: 10.2174/0118715265279242240216114548. Infect Disord Drug Targets. 2024. PMID: 38445691
Cited by
-
Hybrid Immunity and the Incidence of SARS-CoV-2 Reinfections during the Omicron Era in Frontline Healthcare Workers.Vaccines (Basel). 2024 Jun 19;12(6):682. doi: 10.3390/vaccines12060682. Vaccines (Basel). 2024. PMID: 38932411 Free PMC article.
-
Opportunities and challenges encountered in managing cervical cancer during the coronavirus disease 2019 pandemic.Infect Agent Cancer. 2024 Aug 29;19(1):41. doi: 10.1186/s13027-024-00594-3. Infect Agent Cancer. 2024. PMID: 39210452 Free PMC article. Review.
-
The evolution of SARS-CoV-2 seroprevalence in Canada: a time-series study, 2020-2023.CMAJ. 2023 Aug 14;195(31):E1030-E1037. doi: 10.1503/cmaj.230249. CMAJ. 2023. PMID: 37580072 Free PMC article.
References
-
- Bal A, Destras G, Gaymard A, Stefic K, Marlet J, Eymieux S, et al. . Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69-V70, France, August to December 2020. Euro Surveillance. (2021) 26:2100008. 10.2807/1560-7917.ES.2021.26.3.2100008 - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

