Novel CRISPR-based detection of Leishmania species
- PMID: 36187950
- PMCID: PMC9520526
- DOI: 10.3389/fmicb.2022.958693
Novel CRISPR-based detection of Leishmania species
Abstract
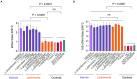
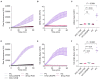
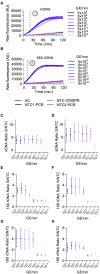
Tegumentary leishmaniasis, a disease caused by protozoan parasites of the genus Leishmania, is a major public health problem in many regions of Latin America. Its diagnosis is difficult given other conditions resembling leishmaniasis lesions and co-occurring in the same endemic areas. A combination of parasitological and molecular methods leads to accurate diagnosis, with the latter being traditionally performed in centralized reference and research laboratories as they require specialized infrastructure and operators. Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein (CRISPR/Cas) systems have recently driven innovative tools for nucleic acid detection that combine high specificity, sensitivity and speed and are readily adaptable for point-of-care testing. Here, we harnessed the CRISPR-Cas12a system for molecular detection of Leishmania spp., emphasizing medically relevant parasite species circulating in Peru and other endemic areas in Latin America, with Leishmania (Viannia) braziliensis being the main etiologic agent of cutaneous and mucosal leishmaniasis. We developed two assays targeting multi-copy targets commonly used in the molecular diagnosis of leishmaniasis: the 18S ribosomal RNA gene (18S rDNA), highly conserved across Leishmania species, and a region of kinetoplast DNA (kDNA) minicircles conserved in the L. (Viannia) subgenus. Our CRISPR-based assays were capable of detecting down to 5 × 10-2 (kDNA) or 5 × 100 (18S rDNA) parasite genome equivalents/reaction with PCR preamplification. The 18S PCR/CRISPR assay achieved pan-Leishmania detection, whereas the kDNA PCR/CRISPR assay was specific for L. (Viannia) detection. No cross-reaction was observed with Trypanosoma cruzi strain Y or human DNA. We evaluated the performance of the assays using 49 clinical samples compared to a kDNA real-time PCR assay as the reference test. The kDNA PCR/CRISPR assay performed equally well as the reference test, with positive and negative percent agreement of 100%. The 18S PCR/CRISPR assay had high positive and negative percent agreement of 82.1% and 100%, respectively. The findings support the potential applicability of the newly developed CRISPR-based molecular tools for first-line diagnosis of Leishmania infections at the genus and L. (Viannia) subgenus levels.
Keywords: 18S rDNA; CRISPR-Cas; Leishmania; invasive and non-invasive clinical specimens; kDNA; molecular diagnostics; nucleic acid detection; tegumentary leishmaniasis.
Copyright © 2022 Dueñas, Nakamoto, Cabrera-Sosa, Huaihua, Cruz, Arévalo, Milón and Adaui.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Adams E. R., Schoone G. J., Ageed A. F., Safi S. E., Schallig H. D. F. H. (2010). Development of a reverse transcriptase loop-mediated isothermal amplification (LAMP) assay for the sensitive detection of Leishmania parasites in clinical samples. Am. J. Trop. Med. Hyg. 82, 591–596. doi: 10.4269/ajtmh.2010.09-0369, PMID: - DOI - PMC - PubMed
-
- Adams E. R., Schoone G., Versteeg I., Gomez M. A., Diro E., Mori Y., et al. . (2018). Development and evaluation of a novel loop-mediated isothermal amplification assay for diagnosis of cutaneous and visceral Leishmaniasis. J. Clin. Microbiol. 56, e00386–18. doi: 10.1128/JCM.00386-18, PMID: - DOI - PMC - PubMed
-
- Adaui V., Lye L.-F., Akopyants N. S., Zimic M., Llanos-Cuentas A., Garcia L., et al. . (2016). Association of the endobiont double-stranded RNA virus LRV1 with treatment failure for human Leishmaniasis caused by Leishmania braziliensis in Peru and Bolivia. J. Infect. Dis. 213, 112–121. doi: 10.1093/infdis/jiv354, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

