T cell receptor repertoire analysis in HTLV-1-associated diseases
- PMID: 36189294
- PMCID: PMC9520328
- DOI: 10.3389/fimmu.2022.984274
T cell receptor repertoire analysis in HTLV-1-associated diseases
Abstract
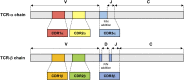
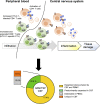
Human T lymphotropic virus 1 (HTLV-1) is a human retrovirus identified as the causative agent in adult T-cell leukemia/lymphoma (ATL) and chronic-progressive neuroinflammatory disorder HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP). HTLV-1 is estimated to infect between 5-20 million people worldwide, although most infected individuals remain asymptomatic. HTLV-1 infected persons carry an estimated lifetime risk of approximately 5% of developing ATL, and between 0.25% and 1.8% of developing HAM/TSP. Most HTLV-1 infection is detected in CD4+ T cells in vivo which causes the aggressive malignancy in ATL. In HAM/TSP, the increase of HTLV-1 provirus induces immune dysregulation to alter inflammatory milieu, such as expansion of HTLV-1-specific CD8+ T cells, in the central nervous system of the infected subjects, which have been suggested to underlie the pathogenesis of HAM/TSP. Factors contributing to the conversion from asymptomatic carrier to disease state remain poorly understood. As such, the identification and tracking of HTLV-1-specific T cell biomarkers that may be used to monitor the progression from primary infection to immune dysfunction and disease are of great interest. T cell receptor (TCR) repertoires have been extensively investigated as a mechanism of monitoring adaptive T cell immune response to viruses and tumors. Breakthrough technologies such as single-cell RNA sequencing have increased the specificity with which T cell clones may be characterized and continue to improve our understanding of TCR signatures in viral infection, cancer, and associated treatments. In HTLV-1-associated disease, sequencing of TCR repertoires has been used to reveal repertoire patterns, diversity, and clonal expansions of HTLV-1-specific T cells capable of immune evasion and dysregulation in ATL as well as in HAM/TSP. Conserved sequence analysis has further been used to identify CDR3 motif sequences and exploit disease- or patient-specificity and commonality in HTLV-1-associated disease. In this article we review current research on TCR repertoires and HTLV-1-specific clonotypes in HTLV-1-associated diseases ATL and HAM/TSP and discuss the implications of TCR clonal expansions on HTLV-1-associated disease course and treatments.
Keywords: HTLV-1-associated myelopathy/tropical spastic paraparesis; T cell receptor (TCR); TCR repertoire; adult T-cell leukemia/lymphoma; human T lymphotropic virus 1.
Copyright © 2022 Clauze, Enose-Akahata and Jacobson.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Identification and tracking of HTLV-1-infected T cell clones in virus-associated neurologic disease.JCI Insight. 2023 Apr 10;8(7):e167422. doi: 10.1172/jci.insight.167422. JCI Insight. 2023. PMID: 37036006 Free PMC article.
-
A Unique T-Cell Receptor Amino Acid Sequence Selected by Human T-Cell Lymphotropic Virus Type 1 Tax301-309-Specific Cytotoxic T Cells in HLA-A24:02-Positive Asymptomatic Carriers and Adult T-Cell Leukemia/Lymphoma Patients.J Virol. 2017 Sep 12;91(19):e00974-17. doi: 10.1128/JVI.00974-17. Print 2017 Oct 1. J Virol. 2017. PMID: 28724766 Free PMC article.
-
Adult T cell leukemia following HTLV-I-associated myelopathy/tropical spastic paraparesis: case reports and implication to the natural course of ATL.Leukemia. 1995 Oct;9(10):1768-70. Leukemia. 1995. PMID: 7564523
-
Tropical spastic paraparesis and HTLV-1 associated myelopathy: clinical, epidemiological, virological and therapeutic aspects.Rev Neurol (Paris). 2012 Mar;168(3):257-69. doi: 10.1016/j.neurol.2011.12.006. Epub 2012 Mar 7. Rev Neurol (Paris). 2012. PMID: 22405461 Review.
-
Immunopathogenesis of HTLV-1-assoaciated myelopathy/tropical spastic paraparesis (HAM/TSP).Life Sci. 2014 May 28;104(1-2):9-14. doi: 10.1016/j.lfs.2014.03.025. Epub 2014 Apr 3. Life Sci. 2014. PMID: 24704970 Review.
Cited by
-
Interferon signaling and ferroptosis in tumor immunology and therapy.NPJ Precis Oncol. 2024 Aug 10;8(1):177. doi: 10.1038/s41698-024-00668-w. NPJ Precis Oncol. 2024. PMID: 39127858 Free PMC article.
-
TLR7 rs179008 (A/T) and TLR7 rs3853839 (C/G) polymorphisms are associated with variations in IFN-α levels in HTLV-1 infection.Front Immunol. 2024 Nov 22;15:1462352. doi: 10.3389/fimmu.2024.1462352. eCollection 2024. Front Immunol. 2024. PMID: 39650644 Free PMC article.
-
Diversity of HLA-A2-Restricted and Immunodominant Epitope Repertoire of Human T-Lymphotropic Virus Type 1 (HTLV-1) Tax Protein: Novel Insights among N-Terminal, Central and C-Terminal Regions.Biomolecules. 2023 Mar 16;13(3):545. doi: 10.3390/biom13030545. Biomolecules. 2023. PMID: 36979478 Free PMC article.
-
Possible Trace of HTLV-1 Virus in Modulation of Cbl-b, ITCH, and PP2A Suppressor Genes.Int J Mol Cell Med. 2025;14(1):472-482. doi: 10.22088/IJMCM.BUMS.14.1.472. Int J Mol Cell Med. 2025. PMID: 40123588 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

