ALIBY: ALFA Nanobody-Based Toolkit for Imaging and Biochemistry in Yeast
- PMID: 36190134
- PMCID: PMC9599267
- DOI: 10.1128/msphere.00333-22
ALIBY: ALFA Nanobody-Based Toolkit for Imaging and Biochemistry in Yeast
Abstract
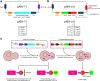
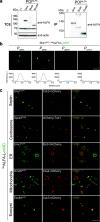
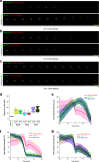
Specialized epitope tags continue to be integral components of various biochemical and cell biological applications such as fluorescence microscopy, immunoblotting, immunoprecipitation, and protein purification. However, until recently, no single tag could offer this complete set of functionalities on its own. Here, we present a plasmid-based toolkit named ALIBY (
Keywords: Saccharomyces cerevisiae; biochemistry; cell biology; cell division; fluorescence; fluorescent image analysis; live-cell imaging; mitochondria; nanobody; vacuoles.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21:947–962. doi: 10.1002/yea.1142. - DOI - PubMed
-
- Götzke H, Kilisch M, Martínez-Carranza M, Sograte-Idrissi S, Rajavel A, Schlichthaerle T, Engels N, Jungmann R, Stenmark P, Opazo F, Frey S. 2019. The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications. Nat Commun 10:4403. doi: 10.1038/s41467-019-12301-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
