The RAC/ROP GTPase activator OsRopGEF10 functions in crown root development by regulating cytokinin signaling in rice
- PMID: 36190337
- PMCID: PMC9806555
- DOI: 10.1093/plcell/koac297
The RAC/ROP GTPase activator OsRopGEF10 functions in crown root development by regulating cytokinin signaling in rice
Abstract
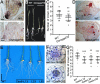
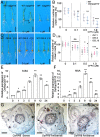
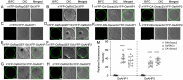
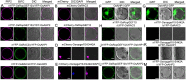
RAC/Rho of plant (ROP) GTPases are major molecular switches that control diverse signaling cascades for plant growth, development, and defense. Here, we discovered a signaling node that connects RAC/ROPs to cytokinins. Rice (Oryza sativa) plants develop a fibrous root system mainly composed of crown roots. Cytokinin signaling via a phosphorelay system is critical for crown root development. We show that OsRopGEF10, which activates RAC/ROPs, acts upstream of the cytoplasmic-nuclear shuttling phosphotransfer proteins AHPs of the cytokinin signaling pathway to promote crown root development. Mutations of OsRopGEF10 induced hypersensitivity to cytokinin, whereas overexpressing this gene reduced the cytokinin response. Loss of OsRopGEF10 function reduced the expression of the response regulator gene OsRR6, a repressor of cytokinin signaling, and impaired crown root development. Mutations in OsAHP1/2 led to increased crown root production and rescued the crown root defect of Osropgef10. Furthermore, auxin activates the ROP GTPase OsRAC3, which attenuates cytokinin signaling for crown root initiation. Molecular interactions between OsRopGEF10, OsRAC3, and OsAHP1/2 implicate a mechanism whereby OsRopGEF10-activated OsRAC3 recruits OsAHP1/2 to the cortical cytoplasm, sequestering them from their phosphorelay function in the nucleus. Together, our findings uncover the OsRopGEF10-OsRAC3-OsAHP1/2 signaling module, establish a link between RAC/ROPs and cytokinin, and reveal molecular crosstalk between auxin and cytokinin during crown root development.
© American Society of Plant Biologists 2022. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures



Similar articles
-
OsRopGEF10 Attenuates Cytokinin Signaling to Regulate Panicle Development and Grain Yield in Rice.Rice (N Y). 2024 Sep 3;17(1):57. doi: 10.1186/s12284-024-00737-5. Rice (N Y). 2024. PMID: 39223425 Free PMC article.
-
The Interaction between Rice ERF3 and WOX11 Promotes Crown Root Development by Regulating Gene Expression Involved in Cytokinin Signaling.Plant Cell. 2015 Sep;27(9):2469-83. doi: 10.1105/tpc.15.00227. Epub 2015 Aug 25. Plant Cell. 2015. PMID: 26307379 Free PMC article.
-
Dynamic cytokinin signaling and function of auxin in cytokinin responsive domains during rice crown root development.Plant Cell Rep. 2021 Aug;40(8):1367-1375. doi: 10.1007/s00299-020-02618-9. Epub 2020 Oct 12. Plant Cell Rep. 2021. PMID: 33047229
-
Sending mixed messages: auxin-cytokinin crosstalk in roots.Curr Opin Plant Biol. 2011 Feb;14(1):10-6. doi: 10.1016/j.pbi.2010.08.014. Curr Opin Plant Biol. 2011. PMID: 20926335 Review.
-
Auxin/Cytokinin Antagonistic Control of the Shoot/Root Growth Ratio and Its Relevance for Adaptation to Drought and Nutrient Deficiency Stresses.Int J Mol Sci. 2022 Feb 9;23(4):1933. doi: 10.3390/ijms23041933. Int J Mol Sci. 2022. PMID: 35216049 Free PMC article. Review.
Cited by
-
The TCP transcription factor TAC8 positively regulates the tiller angle in rice (Oryza sativa L.).Theor Appl Genet. 2025 Jan 30;138(2):39. doi: 10.1007/s00122-024-04812-9. Theor Appl Genet. 2025. PMID: 39885061
-
The TATA-box binding protein-associated factor TAF12b facilitates the degradation of type B response regulators to negatively regulate cytokinin signaling.Plant Commun. 2024 Dec 9;5(12):101076. doi: 10.1016/j.xplc.2024.101076. Epub 2024 Sep 2. Plant Commun. 2024. PMID: 39228128 Free PMC article.
-
Small Papillae Regulated by SPD25 are Critical for Balancing Photosynthetic CO2 Assimilation and Water Loss in Rice.Rice (N Y). 2023 Dec 13;16(1):58. doi: 10.1186/s12284-023-00676-7. Rice (N Y). 2023. PMID: 38087150 Free PMC article.
-
Non-Canonical Inter-Protein Interactions of Key Proteins Belonging to Cytokinin Signaling Pathways.Plants (Basel). 2025 May 15;14(10):1485. doi: 10.3390/plants14101485. Plants (Basel). 2025. PMID: 40431050 Free PMC article. Review.
-
Exploring Hormonal Pathways and Gene Networks in Crown Root Formation Under Stress Conditions: An Update.Plants (Basel). 2025 Feb 19;14(4):630. doi: 10.3390/plants14040630. Plants (Basel). 2025. PMID: 40006889 Free PMC article. Review.
References
-
- Akamatsu A, Wong HL, Fujiwara M, Okuda J, Nishide K, Uno K, Imai K, Umemura K, Kawasaki T, Kawano Y, et al. (2013) An OsCEBiP/OsCERK1–OsRacGEF1–OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 13: 465–476 - PubMed
-
- Argueso CT, Ferreira FJ, Kieber JJ (2009) Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant Cell Environ 32: 1147–1160 - PubMed
-
- Berken A, Wittinghofer A (2008) Structure and function of Rho-type molecular switches in plants. Plant Physiol Biochem 46: 380–393 - PubMed
-
- Berken A, Thomas C, Wittinghofer A (2005) A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 436: 1176–1180 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

