Clinical and Molecular Characterizations of Carbapenem-Resistant Klebsiella pneumoniae Causing Bloodstream Infection in a Chinese Hospital
- PMID: 36190403
- PMCID: PMC9603270
- DOI: 10.1128/spectrum.01690-22
Clinical and Molecular Characterizations of Carbapenem-Resistant Klebsiella pneumoniae Causing Bloodstream Infection in a Chinese Hospital
Abstract
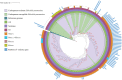
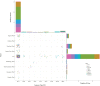
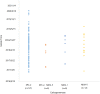
Bloodstream infection (BSI) caused by carbapenem-resistant Klebsiella pneumoniae (CRKP) is a serious and urgent threat for hospitalized patients. This study aims to describe the clinical and molecular characteristics of CRKP causing BSI in a tertiary-care hospital in Beijing, China. A total of 146 CRKP strains and 39 carbapenem-susceptible K. pneumoniae (CSKP) strains collected in the hospital from 2017 to 2020 were sent for whole-genome sequencing. Univariate and multivariate analyses were used to evaluate risk factors for in-hospital mortality of CRKP-BSI cases. Thirty (20.5%) of 146 CRKP-BSI patients and three (7.7%) of 39 CSKP-BSI patients died at discharge (χ2 = 3.471, P = 0.062). Multivariate logistic regression analysis indicated that age and use of urinary catheters were independent risk factors for the death of CRKP-BSI. The 146 CRKP isolates belonged to 9 sequence types (STs) and 11 serotypes, while the 39 CSKP isolates belonged to 23 STs and 27 serotypes. The mechanism of carbapenem resistance for all the CRKP strains was the acquisition of carbapenemase, mainly KPC-2 (n = 127). There were 2 predominant serotypes for ST11 CRKP, namely, KL47 (n = 82) and KL64 (n = 42). Some virulent genes, including rmpA2, iucABCD and iutA, and repB gene, which was involved in plasmid replication, were detected in all ST11-KL64 strains. Evolutionary transmission analysis suggested that ST11 CRKP strains might have evolved from KL47 into KL64 and were accompanied by multiple outbreak events. This study poses an urgent need for enhancing infection control measures in the hospital, especially in the intensive care unit where the patients are at high-risk for acquiring CRKP-BSI. IMPORTANCE CRKP-BSI is demonstrated to cause high mortality. In this study, we demonstrated that ST11 CRKP strains might carry many virulent genes. Meanwhile, outbreak events occurred several times in the strains collected. Carbapenemase acquisition (mainly KPC-2 carbapenemase) was responsible for carbapenem resistance of all the 146 CRKP strains. As 2 predominant strains, all ST11-KL64 strains, but not ST11-KL47 strains, carried rmpA2, iucABCD, iutA, as well as a plasmid replication initiator (repB). Our study suggested that the occurrence of region-specific recombination events manifested by the acquisition of some virulence genes might contribute to serotype switching from ST11-KL47 to ST11-KL64. The accumulation of virulent genes in epidemic resistant strains poses a great challenge for the prevention and treatment of BSI caused by K. pneumoniae in high-risk patients.
Keywords: bloodstream infection; carbapenem resistance; genomic analysis; genomic evolution; transmission.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Genomic Evolution of ST11 Carbapenem-Resistant Klebsiella pneumoniae from 2011 to 2020 Based on Data from the Pathosystems Resource Integration Center.Genes (Basel). 2022 Sep 10;13(9):1624. doi: 10.3390/genes13091624. Genes (Basel). 2022. PMID: 36140792 Free PMC article.
-
Recombination Drives Evolution of Carbapenem-Resistant Klebsiella pneumoniae Sequence Type 11 KL47 to KL64 in China.Microbiol Spectr. 2023 Feb 14;11(1):e0110722. doi: 10.1128/spectrum.01107-22. Epub 2023 Jan 9. Microbiol Spectr. 2023. PMID: 36622219 Free PMC article.
-
Analysis of the virulence of a lethal, carbapenem-resistant hypervirulent KPC-33-producing Klebsiella pneumoniae: Emergence of ST11-KL64 hv-CRKP in ICU.Microb Pathog. 2025 Jan;198:107154. doi: 10.1016/j.micpath.2024.107154. Epub 2024 Nov 23. Microb Pathog. 2025. PMID: 39586335
-
Prevalence and clonal diversity of carbapenem-resistant Klebsiella pneumoniae causing neonatal infections: A systematic review of 128 articles across 30 countries.PLoS Med. 2023 Jun 20;20(6):e1004233. doi: 10.1371/journal.pmed.1004233. eCollection 2023 Jun. PLoS Med. 2023. PMID: 37339120 Free PMC article.
-
Virulence evolution, molecular mechanisms of resistance and prevalence of ST11 carbapenem-resistant Klebsiella pneumoniae in China: A review over the last 10 years.J Glob Antimicrob Resist. 2020 Dec;23:174-180. doi: 10.1016/j.jgar.2020.09.004. Epub 2020 Sep 21. J Glob Antimicrob Resist. 2020. PMID: 32971292 Review.
Cited by
-
A 5-year study of bloodstream infections caused by carbapenemase-producing Gram-negative bacilli in southern Spain.Rev Esp Quimioter. 2024 Dec;37(6):472-478. doi: 10.37201/req/045.2024. Epub 2024 Sep 19. Rev Esp Quimioter. 2024. PMID: 39297392 Free PMC article.
-
Repression of resistance mechanisms of Pseudomonas aeruginosa: implications of the combination of antibiotics and phytoconstituents.Arch Microbiol. 2024 Jun 8;206(7):294. doi: 10.1007/s00203-024-04012-5. Arch Microbiol. 2024. PMID: 38850339 Review.
-
Whole-Genome Sequencing of Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae Isolated from Human Bloodstream Infections.Pathogens. 2025 Feb 20;14(3):205. doi: 10.3390/pathogens14030205. Pathogens. 2025. PMID: 40137690 Free PMC article.
-
Mortality and genetic diversity of antibiotic-resistant bacteria associated with bloodstream infections: a systemic review and genomic analysis.BMC Infect Dis. 2024 Dec 4;24(1):1385. doi: 10.1186/s12879-024-10274-7. BMC Infect Dis. 2024. PMID: 39633294 Free PMC article.
-
Emergence and clinical challenges of ST11-K64 carbapenem-resistant Klebsiella pneumoniae: molecular insights and implications for antimicrobial resistance and virulence in Southwest China.BMC Infect Dis. 2025 Jan 3;25(1):19. doi: 10.1186/s12879-024-10390-4. BMC Infect Dis. 2025. PMID: 39754049 Free PMC article.
References
-
- Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA 112:E3574–E3581. doi:10.1073/pnas.1501049112 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

