HIF2 Inactivation and Tumor Suppression with a Tumor-Directed RNA-Silencing Drug in Mice and Humans
- PMID: 36190432
- PMCID: PMC9771962
- DOI: 10.1158/1078-0432.CCR-22-0963
HIF2 Inactivation and Tumor Suppression with a Tumor-Directed RNA-Silencing Drug in Mice and Humans
Abstract
Purpose: HIF2α is a key driver of kidney cancer. Using a belzutifan analogue (PT2399), we previously showed in tumorgrafts (TG) that ∼50% of clear cell renal cell carcinomas (ccRCC) are HIF2α dependent. However, prolonged treatment induced resistance mutations, which we also identified in humans. Here, we evaluated a tumor-directed, systemically delivered, siRNA drug (siHIF2) active against wild-type and resistant-mutant HIF2α.
Experimental design: Using our credentialed TG platform, we performed pharmacokinetic and pharmacodynamic analyses evaluating uptake, HIF2α silencing, target gene inactivation, and antitumor activity. Orthogonal RNA-sequencing studies of siHIF2 and PT2399 were pursued to define the HIF2 transcriptome. Analyses were extended to a TG line generated from a study biopsy of a siHIF2 phase I clinical trial (NCT04169711) participant and the corresponding patient, an extensively pretreated individual with rapidly progressive ccRCC and paraneoplastic polycythemia likely evidencing a HIF2 dependency.
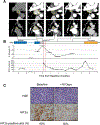
Results: siHIF2 was taken up by ccRCC TGs, effectively depleted HIF2α, deactivated orthogonally defined effector pathways (including Myc and novel E2F pathways), downregulated cell cycle genes, and inhibited tumor growth. Effects on the study subject TG mimicked those in the patient, where HIF2α was silenced in tumor biopsies, circulating erythropoietin was downregulated, polycythemia was suppressed, and a partial response was induced.
Conclusions: To our knowledge, this is the first example of functional inactivation of an oncoprotein and tumor suppression with a systemic, tumor-directed, RNA-silencing drug. These studies provide a proof-of-principle of HIF2α inhibition by RNA-targeting drugs in ccRCC and establish a paradigm for tumor-directed RNA-based therapeutics in cancer.
©2022 American Association for Cancer Research.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

