A draft reference genome of the red abalone, Haliotis rufescens, for conservation genomics
- PMID: 36190478
- PMCID: PMC9709998
- DOI: 10.1093/jhered/esac047
A draft reference genome of the red abalone, Haliotis rufescens, for conservation genomics
Abstract
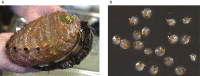
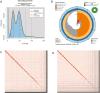
Red abalone, Haliotis rufescens, are herbivorous marine gastropods that primarily feed on kelp. They are the largest and longest-lived of abalone species with a range distribution in North America from central Oregon, United States, to Baja California, MEX. Recently, red abalone have been in decline as a consequence of overharvesting, disease, and climate change, resulting in the closure of the commercial fishery in the 1990s and the recreational fishery in 2018. Protecting this ecologically and economically important species requires an understanding of their current population dynamics and connectivity. Here, we present a new red abalone reference genome as part of the California Conservation Genomics Project (CCGP). Following the CCGP genome strategy, we used Pacific Biosciences HiFi long reads and Dovetail Omni-C data to generate a scaffold-level assembly. The assembly comprises 616 scaffolds for a total size of 1.3 Gb, a scaffold N50 of 45.7 Mb, and a BUSCO complete score of 97.3%. This genome represents a significant improvement over a previous assembly and will serve as a powerful tool for investigating seascape genomic diversity, local adaptation to temperature and ocean acidification, and informing management strategies.
Keywords: CCGP; California Conservation Genomics Project; conservation genomics; red abalone.
© The American Genetic Association. 2022.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

