Conservation at the uterine-placental interface
- PMID: 36191208
- PMCID: PMC9565169
- DOI: 10.1073/pnas.2210633119
Conservation at the uterine-placental interface
Abstract
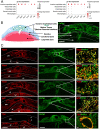
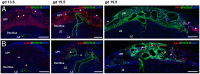
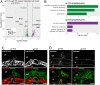
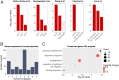
The hemochorial placentation site is characterized by a dynamic interplay between trophoblast cells and maternal cells. These cells cooperate to establish an interface required for nutrient delivery to promote fetal growth. In the human, trophoblast cells penetrate deep into the uterus. This is not a consistent feature of hemochorial placentation and has hindered the establishment of suitable animal models. The rat represents an intriguing model for investigating hemochorial placentation with deep trophoblast cell invasion. In this study, we used single-cell RNA sequencing to characterize the transcriptome of the invasive trophoblast cell lineage, as well as other cell populations within the rat uterine-placental interface during early (gestation day [gd] 15.5) and late (gd 19.5) stages of intrauterine trophoblast cell invasion. We identified a robust set of transcripts that define invasive trophoblast cells, as well as transcripts that distinguished endothelial, smooth muscle, natural killer, and macrophage cells. Invasive trophoblast, immune, and endothelial cell populations exhibited distinct spatial relationships within the uterine-placental interface. Furthermore, the maturation stage of invasive trophoblast cell development could be determined by assessing gestation stage-dependent changes in transcript expression. Finally, and most importantly, expression of a prominent subset of rat invasive trophoblast cell transcripts is conserved in the invasive extravillous trophoblast cell lineage of the human placenta. These findings provide foundational data to identify and interrogate key conserved regulatory mechanisms essential for the development and function of an important compartment within the hemochorial placentation site that is essential for a healthy pregnancy.
Keywords: placentation; rat; single cell genomic analysis; trophoblast.
Conflict of interest statement
The authors declare no competing interest.
Figures

Similar articles
-
TFAP2C is a key regulator of intrauterine trophoblast cell invasion and deep hemochorial placentation.JCI Insight. 2024 Dec 3;10(2):e186471. doi: 10.1172/jci.insight.186471. JCI Insight. 2024. PMID: 39625795 Free PMC article.
-
Intersection of regulatory pathways controlling hemostasis and hemochorial placentation.Proc Natl Acad Sci U S A. 2021 Dec 14;118(50):e2111267118. doi: 10.1073/pnas.2111267118. Proc Natl Acad Sci U S A. 2021. PMID: 34876522 Free PMC article.
-
CITED2 is a conserved regulator of the uterine-placental interface.Proc Natl Acad Sci U S A. 2023 Jan 17;120(3):e2213622120. doi: 10.1073/pnas.2213622120. Epub 2023 Jan 10. Proc Natl Acad Sci U S A. 2023. PMID: 36626551 Free PMC article.
-
Modeling Trophoblast Cell-Guided Uterine Spiral Artery Transformation in the Rat.Int J Mol Sci. 2022 Mar 9;23(6):2947. doi: 10.3390/ijms23062947. Int J Mol Sci. 2022. PMID: 35328368 Free PMC article. Review.
-
Paradigms for investigating invasive trophoblast cell development and contributions to uterine spiral artery remodeling.Placenta. 2021 Sep 15;113:48-56. doi: 10.1016/j.placenta.2021.04.012. Epub 2021 May 3. Placenta. 2021. PMID: 33985793 Free PMC article. Review.
Cited by
-
Cross-Species Insights into Trophoblast Invasion During Placentation Governed by Immune-Featured Trophoblast Cells.Adv Sci (Weinh). 2024 Nov;11(42):e2407221. doi: 10.1002/advs.202407221. Epub 2024 Sep 5. Adv Sci (Weinh). 2024. PMID: 39234818 Free PMC article.
-
NOTUM-MEDIATED WNT SILENCING DRIVES EXTRAVILLOUS TROPHOBLAST CELL LINEAGE DEVELOPMENT.bioRxiv [Preprint]. 2024 Aug 22:2024.02.13.579974. doi: 10.1101/2024.02.13.579974. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2024 Oct;121(40):e2403003121. doi: 10.1073/pnas.2403003121. PMID: 38405745 Free PMC article. Updated. Preprint.
-
TAF7L regulates early stages of male germ cell development in the rat.FASEB J. 2024 Jan;38(1):e23376. doi: 10.1096/fj.202301716RR. FASEB J. 2024. PMID: 38112167 Free PMC article.
-
TFAP2C is a key regulator of intrauterine trophoblast cell invasion and deep hemochorial placentation.JCI Insight. 2024 Dec 3;10(2):e186471. doi: 10.1172/jci.insight.186471. JCI Insight. 2024. PMID: 39625795 Free PMC article.
-
Core conserved transcriptional regulatory networks define the invasive trophoblast cell lineage.Development. 2023 Aug 1;150(15):dev201826. doi: 10.1242/dev.201826. Epub 2023 Jul 31. Development. 2023. PMID: 37417811 Free PMC article.
References
-
- Gardner R. L., Beddington R. S., Multi-lineage ‘stem’ cells in the mammalian embryo. J. Cell Sci. Suppl. 10, 11–27 (1988). - PubMed
-
- Kaufmann P., Black S., Huppertz B., Endovascular trophoblast invasion: Implications for the pathogenesis of intrauterine growth retardation and preeclampsia. Biol. Reprod. 69, 1–7 (2003). - PubMed
-
- Pijnenborg R., Vercruysse L., Hanssens M., The uterine spiral arteries in human pregnancy: Facts and controversies. Placenta 27, 939–958 (2006). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

