NOX1 is essential for TNFα-induced intestinal epithelial ROS secretion and inhibits M cell signatures
- PMID: 36191961
- PMCID: PMC9998338
- DOI: 10.1136/gutjnl-2021-326305
NOX1 is essential for TNFα-induced intestinal epithelial ROS secretion and inhibits M cell signatures
Abstract
Objective: Loss-of-function mutations in genes generating reactive oxygen species (ROS), such as NOX1, are associated with IBD. Mechanisms whereby loss of ROS drive IBD are incompletely defined.
Design: ROS measurements and single-cell transcriptomics were performed on colonoids stratified by NOX1 genotype and TNFα stimulation. Clustering of epithelial cells from human UC (inflamed and uninflamed) scRNASeq was performed. Validation of M cell induction was performed by immunohistochemistry using UEA1 (ulex europaeus agglutin-1 lectin) and in vivo with DSS injury.
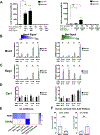
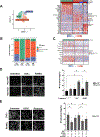
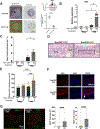
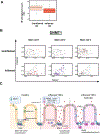
Results: TNFα induces ROS production more in NOX1-WT versus NOX1-deficient murine colonoids under a range of Wnt-mediated and Notch-mediated conditions. scRNASeq from inflamed and uninflamed human colitis versus TNFα stimulated, in vitro colonoids defines substantially shared, induced transcription factors; NOX1-deficient colonoids express substantially lower levels of STAT3 (signal transducer and activator of transcription 3), CEBPD (CCAAT enhancer-binding protein delta), DNMT1 (DNA methyltransferase) and HIF1A (hypoxia-inducible factor) baseline. Subclustering unexpectedly showed marked TNFα-mediated induction of M cells (sentinel cells overlying lymphoid aggregates) in NOX1-deficient colonoids. M cell induction by UEA1 staining is rescued with H2O2 and paraquat, defining extra- and intracellular ROS roles in maintenance of LGR5+ stem cells. DSS injury demonstrated GP2 (glycoprotein-2), basal lymphoplasmacytosis and UEA1 induction in NOX1-deficiency. Principal components analyses of M cell genes and decreased DNMT1 RNA velocity correlate with UC inflammation.
Conclusions: NOX1 deficiency plus TNFα stimulation contribute to colitis through dysregulation of the stem cell niche and altered cell differentiation, enhancing basal lymphoplasmacytosis. Our findings prioritise ROS modulation for future therapies.
Keywords: IBD - genetics; epithelial cells; intestinal stem cell; ulcerative colitis.
© Author(s) (or their employer(s)) 2023. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
