Interleukin-22 regulates neutrophil recruitment in ulcerative colitis and is associated with resistance to ustekinumab therapy
- PMID: 36192482
- PMCID: PMC9530232
- DOI: 10.1038/s41467-022-33331-8
Interleukin-22 regulates neutrophil recruitment in ulcerative colitis and is associated with resistance to ustekinumab therapy
Abstract
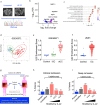
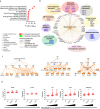
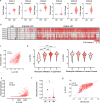
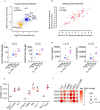
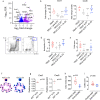
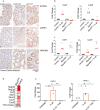
The function of interleukin-22 (IL-22) in intestinal barrier homeostasis remains controversial. Here, we map the transcriptional landscape regulated by IL-22 in human colonic epithelial organoids and evaluate the biological, functional and clinical significance of the IL-22 mediated pathways in ulcerative colitis (UC). We show that IL-22 regulated pro-inflammatory pathways are involved in microbial recognition, cancer and immune cell chemotaxis; most prominently those involving CXCR2+ neutrophils. IL-22-mediated transcriptional regulation of CXC-family neutrophil-active chemokine expression is highly conserved across species, is dependent on STAT3 signaling, and is functionally and pathologically important in the recruitment of CXCR2+ neutrophils into colonic tissue. In UC patients, the magnitude of enrichment of the IL-22 regulated transcripts in colonic biopsies correlates with colonic neutrophil infiltration and is enriched in non-responders to ustekinumab therapy. Our data provide further insights into the biology of IL-22 in human disease and highlight its function in the regulation of pathogenic immune pathways, including neutrophil chemotaxis. The transcriptional networks regulated by IL-22 are functionally and clinically important in UC, impacting patient trajectories and responsiveness to biological intervention.
© 2022. The Author(s).
Conflict of interest statement
K.L., F.Y., A.C.C.S., J.F. were all employed by Janssen Pharmaceuticals. All other authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

