Alternative RNA splicing modulates ribosomal composition and determines the spatial phenotype of glioblastoma cells
- PMID: 36192632
- PMCID: PMC10026424
- DOI: 10.1038/s41556-022-00994-w
Alternative RNA splicing modulates ribosomal composition and determines the spatial phenotype of glioblastoma cells
Abstract
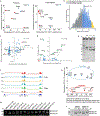
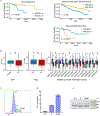
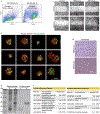
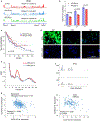
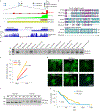
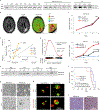
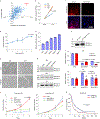
Glioblastoma (GBM) is characterized by exceptionally high intratumoral heterogeneity. However, the molecular mechanisms underlying the origin of different GBM cell populations remain unclear. Here, we found that the compositions of ribosomes of GBM cells in the tumour core and edge differ due to alternative RNA splicing. The acidic pH in the core switches before messenger RNA splicing of the ribosomal gene RPL22L1 towards the RPL22L1b isoform. This allows cells to survive acidosis, increases stemness and correlates with worse patient outcome. Mechanistically, RPL22L1b promotes RNA splicing by interacting with lncMALAT1 in the nucleus and inducing its degradation. Contrarily, in the tumour edge region, RPL22L1a interacts with ribosomes in the cytoplasm and upregulates the translation of multiple messenger RNAs including TP53. We found that the RPL22L1 isoform switch is regulated by SRSF4 and identified a compound that inhibits this process and decreases tumour growth. These findings demonstrate how distinct GBM cell populations arise during tumour growth. Targeting this mechanism may decrease GBM heterogeneity and facilitate therapy.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Figures











Comment in
-
Ribosome specialization in glioblastoma.Nat Cell Biol. 2022 Oct;24(10):1451-1453. doi: 10.1038/s41556-022-01000-z. Nat Cell Biol. 2022. PMID: 36192633 No abstract available.
References
-
- Vaupel P, Kallinowski F & Okunieff P Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 49, 6449–6465 (1989). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

