An evolutionary history of the CoA-binding protein Nat/Ivy
- PMID: 36192822
- PMCID: PMC9703596
- DOI: 10.1002/pro.4463
An evolutionary history of the CoA-binding protein Nat/Ivy
Abstract
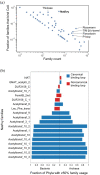
Nat/Ivy is a diverse and ubiquitous CoA-binding evolutionary lineage that catalyzes acyltransferase reactions, primarily converting thioesters into amides. At the heart of the Nat/Ivy fold is a phosphate-binding loop that bears a striking resemblance to that of P-loop NTPases-both are extended, glycine-rich loops situated between a β-strand and an α-helix. Nat/Ivy, therefore, represents an intriguing intersection between thioester chemistry, a putative primitive energy currency, and an ancient mode of phospho-ligand binding. Current evidence suggests that Nat/Ivy emerged independently of other cofactor-utilizing enzymes, and that the observed structural similarity-particularly of the cofactor binding site-is the product of shared constraints instead of shared ancestry. The reliance of Nat/Ivy on a β-α-β motif for CoA-binding highlights the extent to which this simple structural motif may have been a fundamental evolutionary "nucleus" around which modern cofactor-binding domains condensed, as has been suggested for HUP domains, Rossmanns, and P-loop NTPases. Finally, by dissecting the patterns of conserved interactions between Nat/Ivy families and CoA, the coevolution of the enzyme and the cofactor was analyzed. As with the Rossmann, it appears that the pyrophosphate moiety at the center of the cofactor predates the enzyme, suggesting that Nat/Ivy emerged sometime after the metabolite dephospho-CoA.
Keywords: GNAT; P-loop; Rossmann; acetyltransferase; coenzyme A; nucleotide cofactor; phosphate binding loop.
© 2022 The Authors. Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Figures

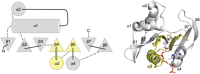
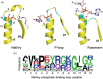




References
-
- Kornprobst M, Turk M, Kellner N, et al. Architecture of the 90S pre‐ribosome: A structural view on the birth of the eukaryotic ribosome. Cell. 2016;166:380–393. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

