Adam10-dependent Notch signaling establishes dental epithelial cell boundaries required for enamel formation
- PMID: 36193048
- PMCID: PMC9526176
- DOI: 10.1016/j.isci.2022.105154
Adam10-dependent Notch signaling establishes dental epithelial cell boundaries required for enamel formation
Abstract
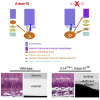
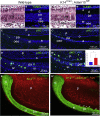
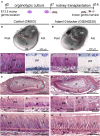
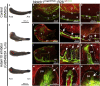
The disintegrin and metalloproteinase Adam10 is a membrane-bound sheddase that regulates Notch signaling and ensures epidermal integrity. To address the function of Adam10 in the continuously growing incisors, we used Keratin14 Cre/+;Adam10 fl/fl transgenic mice, in which Adam10 is conditionally deleted in the dental epithelium. Keratin14 Cre/+;Adam10 fl/fl mice exhibited severe abnormalities, including defective enamel formation reminiscent of human enamel pathologies. Histological analyses of mutant incisors revealed absence of stratum intermedium, and severe disorganization of enamel-secreting ameloblasts. In situ hybridization and immunostaining analyses in the Keratin14 Cre/+;Adam10 fl/fl incisors showed strong Notch1 downregulation in dental epithelium and ectopic distribution of enamel-specific molecules, including ameloblastin and amelogenin. Lineage tracing studies using Notch1 CreERT2 ;R26 mT/mG mice demonstrated that loss of the stratum intermedium cells was due to their fate switch toward the ameloblast lineage. Overall, our data reveal that in the continuously growing incisors the Adam10/Notch axis controls dental epithelial cell boundaries, cell fate switch and proper enamel formation.
Keywords: Cell biology; Developmental biology; Model organism.
© 2022 The Author(s).
Conflict of interest statement
The authors have no conflicts of interest to report.
Figures



Similar articles
-
ADAM10: Possible functions in enamel development.Front Physiol. 2022 Nov 25;13:1032383. doi: 10.3389/fphys.2022.1032383. eCollection 2022. Front Physiol. 2022. PMID: 36505044 Free PMC article. Review.
-
Notch2 Deletion Compromises Epithelial Integrity and Enamel Formation in Rodent Incisors.Cells. 2025 Aug 7;14(15):1224. doi: 10.3390/cells14151224. Cells. 2025. PMID: 40801656 Free PMC article.
-
ADAM10 Expression by Ameloblasts Is Essential for Proper Enamel Formation.Int J Mol Sci. 2024 Dec 7;25(23):13184. doi: 10.3390/ijms252313184. Int J Mol Sci. 2024. PMID: 39684894 Free PMC article.
-
Roles of the histone methyltransferase SET domain bifurcated 1 in epithelial cells during tooth development.Arch Oral Biol. 2024 Sep;165:106026. doi: 10.1016/j.archoralbio.2024.106026. Epub 2024 Jun 10. Arch Oral Biol. 2024. PMID: 38875772
-
Essential roles of ameloblastin in maintaining ameloblast differentiation and enamel formation.Cells Tissues Organs. 2005;181(3-4):189-95. doi: 10.1159/000091380. Cells Tissues Organs. 2005. PMID: 16612084 Review.
Cited by
-
ADAM10: Possible functions in enamel development.Front Physiol. 2022 Nov 25;13:1032383. doi: 10.3389/fphys.2022.1032383. eCollection 2022. Front Physiol. 2022. PMID: 36505044 Free PMC article. Review.
-
Small molecules direct the generation of ameloblast-like cells from human embryonic stem cells.Stem Cell Res Ther. 2025 Apr 12;16(1):173. doi: 10.1186/s13287-025-04294-6. Stem Cell Res Ther. 2025. PMID: 40221796 Free PMC article.
-
Notch Signaling Pathway in Tooth Shape Variations throughout Evolution.Cells. 2023 Feb 27;12(5):761. doi: 10.3390/cells12050761. Cells. 2023. PMID: 36899896 Free PMC article.
-
Adult dental epithelial stem cell-derived organoids deposit hydroxylapatite biomineral.Int J Oral Sci. 2023 Dec 7;15(1):55. doi: 10.1038/s41368-023-00257-w. Int J Oral Sci. 2023. PMID: 38062012 Free PMC article.
-
Notch2 Deletion Compromises Epithelial Integrity and Enamel Formation in Rodent Incisors.Cells. 2025 Aug 7;14(15):1224. doi: 10.3390/cells14151224. Cells. 2025. PMID: 40801656 Free PMC article.
References
-
- Andersson E.R., Sandberg R., Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593–3612. - PubMed
-
- Artavanis-Tsakonas S., Matsuno K., Fortini M.E. Notch signaling. Science. 1995;268:225–232. - PubMed
-
- Artavanis-Tsakonas S., Muskavitch M.A.T. Notch: the past, the present, and the future. Curr. Top. Dev. Biol. 2010;92:1–29. - PubMed
-
- Artavanis-Tsakonas S., Rand M.D., Lake R.J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. - PubMed
-
- Balic A., Thesleff I. Tissue interactions regulating tooth development and renewal. Curr. Top. Dev. Biol. 2015;115:157–186. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

