SARS-CoV-2 3CLpro mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376
- PMID: 36194133
- PMCID: PMC9765458
- DOI: 10.1126/scitranslmed.abq7360
SARS-CoV-2 3CLpro mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376
Abstract
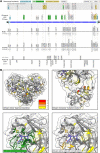
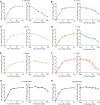
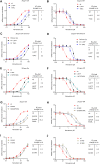
Protease inhibitors are among the most powerful antiviral drugs. Nirmatrelvir is the first protease inhibitor specifically developed against the SARS-CoV-2 protease 3CLpro that has been licensed for clinical use. To identify mutations that confer resistance to this protease inhibitor, we engineered a chimeric vesicular stomatitis virus (VSV) that expressed a polyprotein composed of the VSV glycoprotein (G), the SARS-CoV-2 3CLpro, and the VSV polymerase (L). Viral replication was thus dependent on the autocatalytic processing of this precursor protein by 3CLpro and release of the functional viral proteins G and L, and replication of this chimeric VSV was effectively inhibited by nirmatrelvir. Using this system, we applied nirmatrelvir to select for resistance mutations. Resistance was confirmed by retesting nirmatrelvir against the selected mutations in additional VSV-based systems, in an independently developed cellular system, in a biochemical assay, and in a recombinant SARS-CoV-2 system. We demonstrate that some mutants are cross-resistant to ensitrelvir and GC376, whereas others are less resistant to these compounds. Furthermore, we found that most of these resistance mutations already existed in SARS-CoV-2 sequences that have been deposited in the NCBI and GISAID databases, indicating that these mutations were present in circulating SARS-CoV-2 strains.
Figures


References
-
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). - PMC - PubMed
-
- Voelker R., Combination drug for HCV infection. JAMA 318, 790 (2017). - PubMed
-
- Harrington M., Carpenter C. C. J., Hit HIV-1 hard, but only when necessary. Lancet 355, 2147–2152 (2000). - PubMed
-
- Harcourt B. H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K. M., Smith C. M., Rota P. A., Baker S. C., Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 78, 13600–13612 (2004). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

