Crosstalk between CD4+ T Cells and Airway Smooth Muscle in Pediatric Obesity-related Asthma
- PMID: 36194662
- PMCID: PMC12042998
- DOI: 10.1164/rccm.202205-0985OC
Crosstalk between CD4+ T Cells and Airway Smooth Muscle in Pediatric Obesity-related Asthma
Abstract
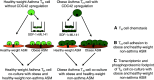
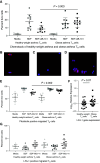
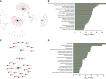
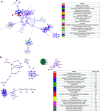
Rationale: Pediatric obesity-related asthma is a nonatopic asthma phenotype with high disease burden and few effective therapies. RhoGTPase upregulation in peripheral blood T helper (Th) cells is associated with the phenotype, but the mechanisms that underlie this association are not known. Objectives: To investigate the mechanisms by which upregulation of CDC42 (Cell Division Cycle 42), a RhoGTPase, in Th cells is associated with airway smooth muscle (ASM) biology. Methods: Chemotaxis of obese asthma and healthy-weight asthma Th cells, and their adhesion to obese and healthy-weight nonasthmatic ASM, was investigated. Transcriptomics and proteomics were used to determine the differential effect of obese and healthy-weight asthma Th cell adhesion to obese or healthy-weight ASM biology. Measurements and Main Results: Chemotaxis of obese asthma Th cells with CDC42 upregulation was resistant to CDC42 inhibition. Obese asthma Th cells were more adherent to obese ASM compared with healthy-weight asthma Th cells to healthy-weight ASM. Compared with coculture with healthy-weight ASM, obese asthma Th cell coculture with obese ASM was positively enriched for genes and proteins involved in actin cytoskeleton organization, transmembrane receptor protein kinase signaling, and cell mitosis, and negatively enriched for extracellular matrix organization. Targeted gene evaluation revealed upregulation of IFNG, TNF (tumor necrosis factor), and Cluster of Differentiation 247 (CD247) among Th cell genes, and of Ak strain transforming (AKT), Ras homolog family member A (RHOA), and CD38, with downregulation of PRKCA (Protein kinase C-alpha), among smooth muscle genes. Conclusions: Obese asthma Th cells have uninhibited chemotaxis and are more adherent to obese ASM, which is associated with upregulation of genes and proteins associated with smooth muscle proliferation and reciprocal nonatopic Th cell activation.
Keywords: Th cells; airway smooth muscle; asthma; children; obesity.
Figures




Comment in
-
Interplay between Immune and Airway Smooth Muscle Cells in Obese Asthma.Am J Respir Crit Care Med. 2023 Feb 15;207(4):388-389. doi: 10.1164/rccm.202210-1870ED. Am J Respir Crit Care Med. 2023. PMID: 36219828 Free PMC article. No abstract available.
References
-
- Rastogi D, Canfield SM, Andrade A, Isasi CR, Hall CB, Rubinstein A, et al. Obesity-associated asthma in children: a distinct entity. Chest . 2012;141:895–905. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

