Myhre syndrome is caused by dominant-negative dysregulation of SMAD4 and other co-factors
- PMID: 36194927
- PMCID: PMC10442510
- DOI: 10.1016/j.diff.2022.09.002
Myhre syndrome is caused by dominant-negative dysregulation of SMAD4 and other co-factors
Abstract
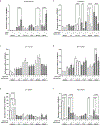
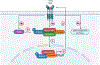
Myhre syndrome is a connective tissue disorder characterized by congenital cardiovascular, craniofacial, respiratory, skeletal, and cutaneous anomalies as well as intellectual disability and progressive fibrosis. It is caused by germline variants in the transcriptional co-regulator SMAD4 that localize at two positions within the SMAD4 protein, I500 and R496, with I500 V/T/M variants more commonly identified in individuals with Myhre syndrome. Here we assess the functional impact of SMAD4-I500V variant, identified in two previously unpublished individuals with Myhre syndrome, and provide novel insights into the molecular mechanism of SMAD4-I500V dysfunction. We show that SMAD4-I500V can dimerize, but its transcriptional activity is severely compromised. Our data show that SMAD4-I500V acts dominant-negatively on SMAD4 and on receptor-regulated SMADs, affecting transcription of target genes. Furthermore, SMAD4-I500V impacts the transcription and function of crucial developmental transcription regulator, NKX2-5. Overall, our data reveal a dominant-negative model of disease for SMAD4-I500V where the function of SMAD4 encoded on the remaining allele, and of co-factors, are perturbed by the continued heterodimerization of the variant, leading to dysregulation of TGF and BMP signaling. Our findings not only provide novel insights into the mechanism of Myhre syndrome pathogenesis but also extend the current knowledge of how pathogenic variants in SMAD proteins cause disease.
Keywords: Dominant-negative; Myhre syndrome; NKX2-5; SMAD4; TGF/BMP signaling; Transcriptional regulation.
Crown Copyright © 2022. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest None.
Figures
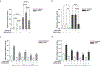


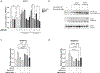
References
-
- Aashaq S, Batool A, Mir SA, Beigh MA, Andrabi KI, Shah ZA (2021) TGF-β signaling: A recap of SMAD-independent and SMAD-dependent pathways. J Cell Physiol - PubMed
-
- Alberici P, Jagmohan-Changur S, De Pater E, Van Der Valk M, Smits R, Hohenstein P, Fodde R (2006) Smad4 haploinsufficiency in mouse models for intestinal cancer. Oncogene 25:1841–1851 - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

