Hydroxymethylation profile of cell-free DNA is a biomarker for early colorectal cancer
- PMID: 36195648
- PMCID: PMC9532421
- DOI: 10.1038/s41598-022-20975-1
Hydroxymethylation profile of cell-free DNA is a biomarker for early colorectal cancer
Abstract
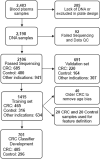
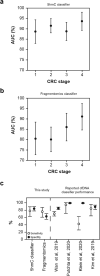
Early detection of cancer will improve survival rates. The blood biomarker 5-hydroxymethylcytosine has been shown to discriminate cancer. In a large covariate-controlled study of over two thousand individual blood samples, we created, tested and explored the properties of a 5-hydroxymethylcytosine-based classifier to detect colorectal cancer (CRC). In an independent validation sample set, the classifier discriminated CRC samples from controls with an area under the receiver operating characteristic curve (AUC) of 90% (95% CI [87, 93]). Sensitivity was 55% at 95% specificity. Performance was similar for early stage 1 (AUC 89%; 95% CI [83, 94]) and late stage 4 CRC (AUC 94%; 95% CI [89, 98]). The classifier could detect CRC even when the proportion of tumor DNA in blood was undetectable by other methods. Expanding the classifier to include information about cell-free DNA fragment size and abundance across the genome led to gains in sensitivity (63% at 95% specificity), with similar overall performance (AUC 91%; 95% CI [89, 94]). We confirm that 5-hydroxymethylcytosine can be used to detect CRC, even in early-stage disease. Therefore, the inclusion of 5-hydroxymethylcytosine in multianalyte testing could improve sensitivity for the detection of early-stage cancer.
© 2022. The Author(s).
Conflict of interest statement
SC, MR, CML, CKL, KH, SM, NJW, LV, MA, MN, TO, HB, SY, JDH, DL, EC and MS are named inventors to patent applications filed by Cambridge Epigenetix Limited pertaining to one or more aspects of the technologies described herein. SY, CKL, DJM, WG, JF, TC, and JDH are current employees of Cambridge Epigenetix Limited. SM, BP, JE, SB and MN have received consulting fees from Cambridge Epigenetix Limited. NJW, MR, SY, CKL, CML, KH, DJM, WG, JF, TC, HB, GR, SB, SC, ML, YC, CJ, LK, MMA, EC, VW, LM, TB, ATF, NS, PG, MK, SM, LV, SS, MA, DB, MJ, DL, JM, MS, TO, VP, MM, DS, AV, SB and JDH have rights to share options in Cambridge Epigenetix Limited.
Figures



References
-
- Kakushadze Z, Raghubanshi R, Yu W. Estimating cost savings from early cancer diagnosis. Data. 2017 doi: 10.3390/data2030030. - DOI

