Higher phage virulence accelerates the evolution of host resistance
- PMID: 36196537
- PMCID: PMC9532999
- DOI: 10.1098/rspb.2022.1070
Higher phage virulence accelerates the evolution of host resistance
Abstract
Pathogens vary strikingly in their virulence and the selection they impose on their hosts. While the evolution of different virulence levels is well studied, the evolution of host resistance in response to different virulence levels is less understood and, at present, mainly based on observations and theoretical predictions with few experimental tests. Increased virulence can increase selection for host resistance evolution if the benefits of avoiding infection outweigh resistance costs. To test this, we experimentally evolved the bacterium Vibrio alginolyticus in the presence of two variants of a filamentous phage that differ in their virulence. The bacterial host exhibited two alternative defence strategies: (1) super infection exclusion (SIE), whereby phage-infected cells were immune to subsequent infection at the cost of reduced growth, and (2) surface receptor mutations (SRM), providing resistance to infection by preventing phage attachment. While SIE emerged rapidly against both phages, SRM evolved faster against the high- than the low-virulence phage. Using a mathematical model of our system, we show that increasing virulence strengthens selection for SRM owing to the higher costs of infection suffered by SIE immune hosts. Thus, by accelerating the evolution of host resistance, more virulent phages caused shorter epidemics.
Keywords: experimental evolution; filamentous phages; resistance evolution; virulence.
Conflict of interest statement
We declare we have no competing interests.
Figures
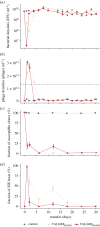


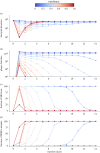
References
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

