SYNJ2BP Improves the Production of Lentiviral Envelope Protein by Facilitating the Formation of Mitochondrion-Associated Endoplasmic Reticulum Membrane
- PMID: 36197105
- PMCID: PMC9599250
- DOI: 10.1128/jvi.00549-22
SYNJ2BP Improves the Production of Lentiviral Envelope Protein by Facilitating the Formation of Mitochondrion-Associated Endoplasmic Reticulum Membrane
Abstract
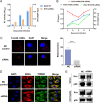
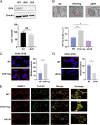
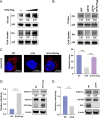
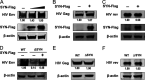
Equine infectious anemia virus (EIAV) and HIV are both members of the Lentivirus genus and are similar in major virological characters. EIAV endangers the horse industry. In addition, EIAV can also be used as a model for HIV research. The maturation of the lentiviral Env protein, which is necessary for viral entry, requires Env to be folded in the endoplasmic reticulum (ER). It is currently unclear how this process is regulated. Mitochondrion-associated endoplasmic reticulum membrane (MAM) is a specialized part of the close connection between the ER and mitochondria, and one of the main functions of MAM is to promote oxidative protein production in the ER. SYNJ2BP is one of the key proteins that make up the MAM, and we found that SYNJ2BP is essential for EIAV replication. We therefore constructed a SYNJ2BP knockout HEK293T cell line in which the number of MAMs is significantly reduced. Moreover, overexpression of SYNJ2BP could increase the number of MAMs. Our study demonstrates that SYNJ2BP can improve the infectivity of the EIAV virus with elevated production of the viral Env protein through increased MAM formation. Interestingly, SYNJ2BP was able to improve the production of not only EIAV Env but also HIV. Further investigation showed that MAMs can provide more ATP and calcium ions, which are essential factors for Env production, to the ER and can also reduce ER stress induced by HIV or EIAV Envs to increase the Env production level in cells. These results may help us to understand the key production mechanisms of lentiviral Env. IMPORTANCE Lentiviral Env proteins, which are rich in disulfide bonds, need to be fully folded in the ER; otherwise, misfolded Env proteins will induce ER stress and be degraded by ER-associated protein degradation (ERAD). To date, it is still unclear about Env production mechanism in the ER. MAM is the structure of closely connection between the ER and mitochondria. MAMs play important roles in the calcium steady state and oxidative stress, especially in the production of oxidative protein. For the first time, we found that SYNJ2BP can promote the production of lentiviral Env proteins by providing the ATP and calcium ions required for oxidative protein production in the ER and by reducing ER stress through facilitating formation of MAMs. These studies shed light on how MAMs improve lentiviral Env production, which will lay the foundation for the study of replication mechanisms in other lentiviruses from the perspective of the cellular organelle microenvironment.
Keywords: EIAV; HIV; SYNJ2BP; endoplasmic reticulum; envelope protein; lentivirus; mitochondria; mitochondrion-associated endoplasmic reticulum membrane.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Truncation of the Cytoplasmic Tail of Equine Infectious Anemia Virus Increases Virion Production by Improving Env Cleavage and Plasma Membrane Localization.J Virol. 2021 Nov 9;95(23):e0108721. doi: 10.1128/JVI.01087-21. Epub 2021 Sep 8. J Virol. 2021. PMID: 34495693 Free PMC article.
-
Equine viperin restricts equine infectious anemia virus replication by inhibiting the production and/or release of viral Gag, Env, and receptor via distortion of the endoplasmic reticulum.J Virol. 2014 Nov;88(21):12296-310. doi: 10.1128/JVI.01379-14. Epub 2014 Aug 13. J Virol. 2014. PMID: 25122784 Free PMC article.
-
The mitochondrial translocator protein, TSPO, inhibits HIV-1 envelope glycoprotein biosynthesis via the endoplasmic reticulum-associated protein degradation pathway.J Virol. 2014 Mar;88(6):3474-84. doi: 10.1128/JVI.03286-13. Epub 2014 Jan 8. J Virol. 2014. PMID: 24403586 Free PMC article.
-
The Molecular Mechanisms Underlying Mitochondria-Associated Endoplasmic Reticulum Membrane-Induced Insulin Resistance.Front Endocrinol (Lausanne). 2020 Nov 23;11:592129. doi: 10.3389/fendo.2020.592129. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 33329397 Free PMC article. Review.
-
Oxidative protein folding in the endoplasmic reticulum: tight links to the mitochondria-associated membrane (MAM).Biochim Biophys Acta. 2010 Aug;1798(8):1465-73. doi: 10.1016/j.bbamem.2010.04.009. Epub 2010 Apr 27. Biochim Biophys Acta. 2010. PMID: 20430008 Free PMC article. Review.
Cited by
-
ESYT1 tethers the ER to mitochondria and is required for mitochondrial lipid and calcium homeostasis.Life Sci Alliance. 2023 Nov 6;7(1):e202302335. doi: 10.26508/lsa.202302335. Print 2024 Jan. Life Sci Alliance. 2023. PMID: 37931956 Free PMC article.
-
Viral regulation of organelle membrane contact sites.PLoS Biol. 2024 Mar 5;22(3):e3002529. doi: 10.1371/journal.pbio.3002529. eCollection 2024 Mar. PLoS Biol. 2024. PMID: 38442090 Free PMC article.
-
Nitazoxanide controls virus viability through its impact on membrane bioenergetics.Sci Rep. 2024 Dec 28;14(1):30679. doi: 10.1038/s41598-024-78694-8. Sci Rep. 2024. PMID: 39730386 Free PMC article.
-
Dengue virus and Zika virus alter endoplasmic reticulum-mitochondria contact sites to regulate respiration and apoptosis.iScience. 2024 Dec 14;28(1):111599. doi: 10.1016/j.isci.2024.111599. eCollection 2025 Jan 17. iScience. 2024. PMID: 39834870 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

