A Small Molecule Reacts with the p53 Somatic Mutant Y220C to Rescue Wild-type Thermal Stability
- PMID: 36197521
- PMCID: PMC9827106
- DOI: 10.1158/2159-8290.CD-22-0381
A Small Molecule Reacts with the p53 Somatic Mutant Y220C to Rescue Wild-type Thermal Stability
Abstract
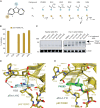
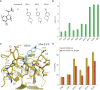
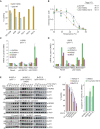
The transcription factor and tumor suppressor protein p53 is the most frequently mutated and inactivated gene in cancer. Mutations in p53 result in deregulated cell proliferation and genomic instability, both hallmarks of cancer. There are currently no therapies available that directly target mutant p53 to rescue wild-type function. In this study, we identify covalent compsounds that selectively react with the p53 somatic mutant cysteine Y220C and restore wild-type thermal stability.
Significance: The tumor suppressor p53 is the most mutated gene in cancer, and yet no therapeutics to date directly target the mutated protein to rescue wild-type function. In this study, we identify the first allele-specific compound that selectively reacts with the cysteine p53 Y220C to rescue wild-type thermal stability and gene activation. See related commentary by Lane and Verma, p. 14. This article is highlighted in the In This Issue feature, p. 1.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures

Comment in
-
Covalent Rescue of Mutant p53.Cancer Discov. 2023 Jan 9;13(1):14-16. doi: 10.1158/2159-8290.CD-22-1212. Cancer Discov. 2023. PMID: 36620883
References
-
- McBride KA, Ballinger ML, Killick E, Kirk J, Tattersall MHN, Eeles RA, et al. . Li-Fraumeni syndrome: cancer risk assessment and clinical management. Nat Rev Clin Oncol 2014;11:260–71. - PubMed
-
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. . Restoration of p53 function leads to tumour regression in vivo. Nature 2007;445:661–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

