The RNA helicase DDX6 controls early mouse embryogenesis by repressing aberrant inhibition of BMP signaling through miRNA-mediated gene silencing
- PMID: 36197846
- PMCID: PMC9534413
- DOI: 10.1371/journal.pgen.1009967
The RNA helicase DDX6 controls early mouse embryogenesis by repressing aberrant inhibition of BMP signaling through miRNA-mediated gene silencing
Abstract
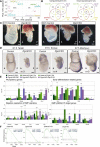
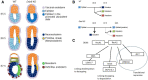
The evolutionarily conserved RNA helicase DDX6 is a central player in post-transcriptional regulation, but its role during embryogenesis remains elusive. We here show that DDX6 enables proper cell lineage specification from pluripotent cells by analyzing Ddx6 knockout (KO) mouse embryos and employing an in vitro epiblast-like cell (EpiLC) induction system. Our study unveils that DDX6 is an important BMP signaling regulator. Deletion of Ddx6 causes the aberrant upregulation of the negative regulators of BMP signaling, which is accompanied by enhanced expression of Nodal and related genes. Ddx6 KO pluripotent cells acquire higher pluripotency with a strong inclination toward neural lineage commitment. During gastrulation, abnormally expanded Nodal and Eomes expression in the primitive streak likely promotes endoderm cell fate specification while inhibiting mesoderm differentiation. We also genetically dissected major DDX6 pathways by generating Dgcr8, Dcp2, and Eif4enif1 KO models in addition to Ddx6 KO. We found that the miRNA pathway mutant Dgcr8 KO phenocopies Ddx6 KO, indicating that DDX6 mostly works along with the miRNA pathway during early development, whereas its P-body-related functions are dispensable. Therefore, we conclude that DDX6 prevents aberrant upregulation of BMP signaling inhibitors by participating in miRNA-mediated gene silencing processes. Overall, this study delineates how DDX6 affects the development of the three primary germ layers during early mouse embryogenesis and the underlying mechanism of DDX6 function.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

