Recent Advances in Biosensors for Detection of COVID-19 and Other Viruses
- PMID: 36197867
- PMCID: PMC10009816
- DOI: 10.1109/RBME.2022.3212038
Recent Advances in Biosensors for Detection of COVID-19 and Other Viruses
Abstract
This century has introduced very deadly, dangerous, and infectious diseases to humankind such as the influenza virus, Ebola virus, Zika virus, and the most infectious SARS-CoV-2 commonly known as COVID-19 and have caused epidemics and pandemics across the globe. For some of these diseases, proper medications, and vaccinations are missing and the early detection of these viruses will be critical to saving the patients. And even the vaccines are available for COVID-19, the new variants of COVID-19 such as Delta, and Omicron are spreading at large. The available virus detection techniques take a long time, are costly, and complex and some of them generates false negative or false positive that might cost patients their lives. The biosensor technique is one of the best qualified to address this difficult challenge. In this systematic review, we have summarized recent advancements in biosensor-based detection of these pandemic viruses including COVID-19. Biosensors are emerging as efficient and economical analytical diagnostic instruments for early-stage illness detection. They are highly suitable for applications related to healthcare, wearable electronics, safety, environment, military, and agriculture. We strongly believe that these insights will aid in the study and development of a new generation of adaptable virus biosensors for fellow researchers.
Figures
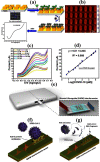


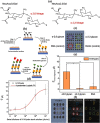

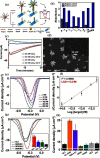
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

