A non-canonical retina-ipRGCs-SCN-PVT visual pathway for mediating contagious itch behavior
- PMID: 36198265
- PMCID: PMC9595067
- DOI: 10.1016/j.celrep.2022.111444
A non-canonical retina-ipRGCs-SCN-PVT visual pathway for mediating contagious itch behavior
Abstract
Contagious itch behavior informs conspecifics of adverse environment and is crucial for the survival of social animals. Gastrin-releasing peptide (GRP) and its receptor (GRPR) in the suprachiasmatic nucleus (SCN) of the hypothalamus mediates contagious itch behavior in mice. Here, we show that intrinsically photosensitive retina ganglion cells (ipRGCs) convey visual itch information, independently of melanopsin, from the retina to GRP neurons via PACAP-PAC1R signaling. Moreover, GRPR neurons relay itch information to the paraventricular nucleus of the thalamus (PVT). Surprisingly, neither the visual cortex nor superior colliculus is involved in contagious itch. In vivo calcium imaging and extracellular recordings reveal contagious itch-specific neural dynamics of GRPR neurons. Thus, we propose that the retina-ipRGC-SCN-PVT pathway constitutes a previously unknown visual pathway that probably evolved for motion vision that encodes salient environmental cues and enables animals to imitate behaviors of conspecifics as an anticipatory mechanism to cope with adverse conditions.
Keywords: CP: Neuroscience; GRP; GRPR; PACAP; SCN; contagious itch behavior; ipRGCs; itch stimuli; neural dynamics; scratching motion; stress.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

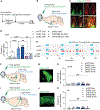

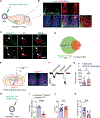
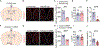
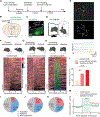

References
-
- Abrahamson EE, and Moore RY (2001). Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 916, 172–191. - PubMed
-
- Baver SB, Pickard GE, Sollars PJ, and Pickard GE (2008). Two types of melanopsin retinal ganglion cell differentially innervate the hypothalamic suprachiasmatic nucleus and the olivary pretectal nucleus. Eur. J. Neurosci. 27, 1763–1770. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

