Temperature synchronisation of circadian rhythms in primary human airway epithelial cells from children
- PMID: 36198442
- PMCID: PMC9535174
- DOI: 10.1136/bmjresp-2022-001319
Temperature synchronisation of circadian rhythms in primary human airway epithelial cells from children
Abstract
Introduction: Cellular circadian rhythms regulate immune pathways and inflammatory responses that mediate human disease such as asthma. Circadian rhythms in the lung may also contribute to exacerbations of chronic diseases such as asthma by regulating observed rhythms in mucus production, bronchial reactivity, airway inflammation and airway resistance. Primary human airway epithelial cells (AECs) are commonly used to model human lung diseases, such as asthma, with circadian symptoms, but a method for synchronising circadian rhythms in AECs has not been developed, and the presence of circadian rhythms in human AECs remains uninvestigated.
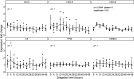
Methods: We used temperature cycling to synchronise circadian rhythms in undifferentiated and differentiated primary human AECs. Reverse transcriptase-quantitative PCR was used to measure expression of the core circadian clock genes ARNTL, CLOCK, CRY1, CRY2, NR1D1, NR1D2, PER1 and PER2.
Results: Following temperature synchronisation, the core circadian genes ARNTL, CRY1, CRY2, NR1D1, NR1D2, PER1 and PER2 maintained endogenous 24-hour rhythms under constant conditions. Following serum shock, the core circadian genes ARNTL, NR1D1 and NR1D2 demonstrated rhythmic expression. Following temperature synchronisation, CXCL8 demonstrated rhythmic circadian expression.
Conclusions: Temperature synchronised circadian rhythms in AECs differentiated at an air-liquid interface can serve as a model to investigate circadian rhythms in pulmonary diseases.
Keywords: airway epithelium; asthma mechanisms; paediatric asthma; paediatric lung disaese; respiratory infection; sleep apnoea.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
