STING-induced regulatory B cells compromise NK function in cancer immunity
- PMID: 36198789
- PMCID: PMC9875944
- DOI: 10.1038/s41586-022-05254-3
STING-induced regulatory B cells compromise NK function in cancer immunity
Abstract
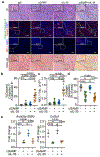
An immunosuppressive tumour microenvironment is a major obstacle in the control of pancreatic and other solid cancers1-3. Agonists of the stimulator of interferon genes (STING) protein trigger inflammatory innate immune responses to potentially overcome tumour immunosuppression4. Although these agonists hold promise as potential cancer therapies5, tumour resistance to STING monotherapy has emerged in clinical trials and the mechanism(s) is unclear5-7. Here we show that the administration of five distinct STING agonists, including cGAMP, results in an expansion of human and mouse interleukin (IL)-35+ regulatory B cells in pancreatic cancer. Mechanistically, cGAMP drives expression of IL-35 by B cells in an IRF3-dependent but type I interferon-independent manner. In several preclinical cancer models, the loss of STING signalling in B cells increases tumour control. Furthermore, anti-IL-35 blockade or genetic ablation of IL-35 in B cells also reduces tumour growth. Unexpectedly, the STING-IL-35 axis in B cells reduces proliferation of natural killer (NK) cells and attenuates the NK-driven anti-tumour response. These findings reveal an intrinsic barrier to systemic STING agonist monotherapy and provide a combinatorial strategy to overcome immunosuppression in tumours.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Conflict of Interest
The authors have declared that no conflict of interest exists.
Figures






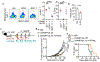







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

