Collagenolysis-dependent DDR1 signalling dictates pancreatic cancer outcome
- PMID: 36198801
- PMCID: PMC9588640
- DOI: 10.1038/s41586-022-05169-z
Collagenolysis-dependent DDR1 signalling dictates pancreatic cancer outcome
Erratum in
-
Publisher Correction: Collagenolysis-dependent DDR1 signalling dictates pancreatic cancer outcome.Nature. 2023 Mar;615(7953):E24. doi: 10.1038/s41586-023-05920-0. Nature. 2023. PMID: 36882540 Free PMC article. No abstract available.
-
Author Correction: Collagenolysis-dependent DDR1 signalling dictates pancreatic cancer outcome.Nature. 2025 Jul;643(8071):E10. doi: 10.1038/s41586-025-09285-4. Nature. 2025. PMID: 40533566 Free PMC article. No abstract available.
Abstract
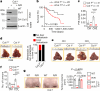
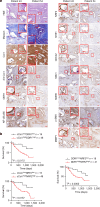
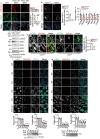
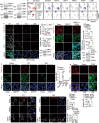
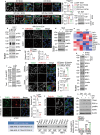
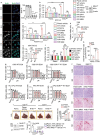
Pancreatic ductal adenocarcinoma (PDAC) is a highly desmoplastic, aggressive cancer that frequently progresses and spreads by metastasis to the liver1. Cancer-associated fibroblasts, the extracellular matrix and type I collagen (Col I) support2,3 or restrain the progression of PDAC and may impede blood supply and nutrient availability4. The dichotomous role of the stroma in PDAC, and the mechanisms through which it influences patient survival and enables desmoplastic cancers to escape nutrient limitation, remain poorly understood. Here we show that matrix-metalloprotease-cleaved Col I (cCol I) and intact Col I (iCol I) exert opposing effects on PDAC bioenergetics, macropinocytosis, tumour growth and metastasis. Whereas cCol I activates discoidin domain receptor 1 (DDR1)-NF-κB-p62-NRF2 signalling to promote the growth of PDAC, iCol I triggers the degradation of DDR1 and restrains the growth of PDAC. Patients whose tumours are enriched for iCol I and express low levels of DDR1 and NRF2 have improved median survival compared to those whose tumours have high levels of cCol I, DDR1 and NRF2. Inhibition of the DDR1-stimulated expression of NF-κB or mitochondrial biogenesis blocks tumorigenesis in wild-type mice, but not in mice that express MMP-resistant Col I. The diverse effects of the tumour stroma on the growth and metastasis of PDAC and on the survival of patients are mediated through the Col I-DDR1-NF-κB-NRF2 mitochondrial biogenesis pathway, and targeting components of this pathway could provide therapeutic opportunities.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures










References
-
- Erkan, M. et al. The activated stroma index is a novel and independent prognostic marker in pancreatic ductal adenocarcinoma. Clin. Gastroenterol. Hepatol.6, 1155–1161 (2008). - PubMed
MeSH terms
Substances
Grants and funding
- R01 CA218254/CA/NCI NIH HHS/United States
- R01 CA234245/CA/NCI NIH HHS/United States
- R37 AI043477/AI/NIAID NIH HHS/United States
- P01 DK098108/DK/NIDDK NIH HHS/United States
- P30 NS047101/NS/NINDS NIH HHS/United States
- R01 CA190844/CA/NCI NIH HHS/United States
- P30 CA023100/CA/NCI NIH HHS/United States
- U01 AA027681/AA/NIAAA NIH HHS/United States
- P30 CA014195/CA/NCI NIH HHS/United States
- P30 DK132710/DK/NIDDK NIH HHS/United States
- R01 CA211794/CA/NCI NIH HHS/United States
- U01 CA274295/CA/NCI NIH HHS/United States
- R01 CA262424/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

