System analysis based on the pyroptosis-related genes identifies GSDMC as a novel therapy target for pancreatic adenocarcinoma
- PMID: 36199146
- PMCID: PMC9533512
- DOI: 10.1186/s12967-022-03632-z
System analysis based on the pyroptosis-related genes identifies GSDMC as a novel therapy target for pancreatic adenocarcinoma
Abstract
Background: Pancreatic adenocarcinoma (PAAD) is one of the most common malignant tumors of the digestive tract. Pyroptosis is a newly discovered programmed cell death that highly correlated with the prognosis of tumors. However, the prognostic value of pyroptosis in PAAD remains unclear.
Methods: A total of 178 pancreatic cancer PAAD samples and 167 normal samples were obtained from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases. The "DESeq2" R package was used to identify differntially expressed pyroptosis-related genes between normal pancreatic samples and PAAD samples. The prognostic model was established in TCGA cohort based on univariate Cox and the least absolute shrinkage and selection operator (LASSO) Cox regression analyses, which was validated in test set from Gene Expression Omnibus (GEO) cohort. Univariate independent prognostic analysis and multivariate independent prognostic analysis were used to determine whether the risk score can be used as an independent prognostic factor to predict the clinicopathological features of PAAD patients. A nomogram was used to predict the survival probability of PAAD patients, which could help in clinical decision-making. The R package "pRRophetic" was applied to calculate the drug sensitivity of each samples from high- and low-risk group. Tumor immune infiltration was investigated using an ESTIMATE algorithm. Finally, the pro-tumor phenotype of GSDMC was explored in PANC-1 and CFPAC-1 cells.
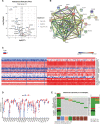
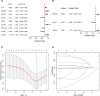
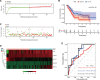
Result: On the basis of univariate Cox and LASSO regression analyses, we constructed a risk model with identified five pyroptosis-related genes (IL18, CASP4, NLRP1, GSDMC, and NLRP2), which was validated in the test set. The PAAD samples were divided into high-risk and low-risk groups on the basis of the risk score's median. According to Kaplan Meier curve analysis, samples from high-risk groups had worse outcomes than those from low-risk groups. The time-dependent receiver operating characteristics (ROC) analysis revealed that the risk model could predict the prognosis of PAAD accurately. A nomogram accompanied by calibration curves was presented for predicting 1-, 2-, and 3-year survival in PAAD patients. More importantly, 4 small molecular compounds (A.443654, PD.173074, Epothilone. B, Lapatinib) were identified, which might be potential drugs for the treatment of PAAD patients. Finally, the depletion of GSDMC inhibits the proliferation, invasion, and migration of pancreatic adenocarcinoma cells.
Conclusion: In this study, we developed a pyroptosis-related prognostic model based on IL18, CASP4, NLRP1, NLRP2, and GSDMC , which may be helpful for clinicians to make clinical decisions for PAAD patients and provide valuable insights for individualized treatment. Our result suggest that GSDMC may promote the proliferation and migration of PAAD cell lines. These findings may provide new insights into the roles of pyroptosis-related genes in PAAD, and offer new therapeutic targets for the treatment of PAAD.
Keywords: Drug; GSDMC; Immune infiltration; Pancreatic adenocarcinoma; Prognostic model; Pyroptosis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Development of a Prognostic Model Based on Pyroptosis-Related Genes in Pancreatic Adenocarcinoma.Dis Markers. 2022 May 29;2022:9141117. doi: 10.1155/2022/9141117. eCollection 2022. Dis Markers. 2022. PMID: 35677632 Free PMC article.
-
System analysis based on the pyroptosis-related genes identifes GSDMD as a novel therapy target for skin cutaneous melanoma.J Transl Med. 2023 Nov 10;21(1):801. doi: 10.1186/s12967-023-04513-9. J Transl Med. 2023. PMID: 37950289 Free PMC article.
-
Development and Validation of an Inflammatory Response-Related Gene Signature for Predicting the Prognosis of Pancreatic Adenocarcinoma.Inflammation. 2022 Aug;45(4):1732-1751. doi: 10.1007/s10753-022-01657-6. Epub 2022 Mar 23. Inflammation. 2022. PMID: 35322324
-
Identify potential prognostic indicators and tumor-infiltrating immune cells in pancreatic adenocarcinoma.Biosci Rep. 2022 Feb 25;42(2):BSR20212523. doi: 10.1042/BSR20212523. Biosci Rep. 2022. PMID: 35083488 Free PMC article. Review.
-
Pyroptosis: mechanisms and diseases.Signal Transduct Target Ther. 2021 Mar 29;6(1):128. doi: 10.1038/s41392-021-00507-5. Signal Transduct Target Ther. 2021. PMID: 33776057 Free PMC article. Review.
Cited by
-
Role of gasdermin family proteins in cancers (Review).Int J Oncol. 2023 Sep;63(3):100. doi: 10.3892/ijo.2023.5548. Epub 2023 Jul 21. Int J Oncol. 2023. PMID: 37477150 Free PMC article.
-
Diagnostic potential of CDK1 and STAT1 in acute kidney injury associated with gastrointestinal cancers: a bioinformatics-based study.Front Mol Biosci. 2025 Jan 22;12:1522246. doi: 10.3389/fmolb.2025.1522246. eCollection 2025. Front Mol Biosci. 2025. PMID: 39911265 Free PMC article.
-
Construction and comprehensive analysis of a novel prognostic signature associated with pyroptosis molecular subtypes in patients with pancreatic adenocarcinoma.Front Immunol. 2023 Feb 3;14:1111494. doi: 10.3389/fimmu.2023.1111494. eCollection 2023. Front Immunol. 2023. PMID: 36817451 Free PMC article.
-
Liquid-liquid phase separation-related features of PYGB/ACTR3/CCNA2/ITGB1/ATP8A1/RAP1GAP2 predict the prognosis of pancreatic cancer.J Gastrointest Oncol. 2024 Aug 31;15(4):1723-1745. doi: 10.21037/jgo-24-426. Epub 2024 Aug 22. J Gastrointest Oncol. 2024. PMID: 39279964 Free PMC article.
-
The expression profile of Gasdermin C-related genes predicts the prognosis and immunotherapy response of pancreatic adenocarcinoma.Am J Cancer Res. 2023 Apr 15;13(4):1240-1258. eCollection 2023. Am J Cancer Res. 2023. PMID: 37168356 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

