Microbiome-driven breeding strategy potentially improves beef fatty acid profile benefiting human health and reduces methane emissions
- PMID: 36199148
- PMCID: PMC9533493
- DOI: 10.1186/s40168-022-01352-6
Microbiome-driven breeding strategy potentially improves beef fatty acid profile benefiting human health and reduces methane emissions
Erratum in
-
Correction: Microbiome-driven breeding strategy potentially improves beef fatty acid profile benefiting human health and reduces methane emissions.Microbiome. 2022 Oct 25;10(1):184. doi: 10.1186/s40168-022-01392-y. Microbiome. 2022. PMID: 36280892 Free PMC article. No abstract available.
Abstract
Background: Healthier ruminant products can be achieved by adequate manipulation of the rumen microbiota to increase the flux of beneficial fatty acids reaching host tissues. Genomic selection to modify the microbiome function provides a permanent and accumulative solution, which may have also favourable consequences in other traits of interest (e.g. methane emissions). Possibly due to a lack of data, this strategy has never been explored.
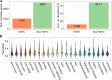
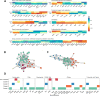
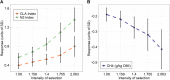
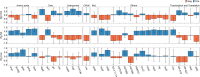
Results: This study provides a comprehensive identification of ruminal microbial mechanisms under host genomic influence that directly or indirectly affect the content of unsaturated fatty acids in beef associated with human dietary health benefits C18:3n-3, C20:5n-3, C22:5n-3, C22:6n-3 or cis-9, trans-11 C18:2 and trans-11 C18:1 in relation to hypercholesterolemic saturated fatty acids C12:0, C14:0 and C16:0, referred to as N3 and CLA indices. We first identified that ~27.6% (1002/3633) of the functional core additive log-ratio transformed microbial gene abundances (alr-MG) in the rumen were at least moderately host-genomically influenced (HGFC). Of these, 372 alr-MG were host-genomically correlated with the N3 index (n=290), CLA index (n=66) or with both (n=16), indicating that the HGFC influence on beef fatty acid composition is much more complex than the direct regulation of microbial lipolysis and biohydrogenation of dietary lipids and that N3 index variation is more strongly subjected to variations in the HGFC than CLA. Of these 372 alr-MG, 110 were correlated with the N3 and/or CLA index in the same direction, suggesting the opportunity for enhancement of both indices simultaneously through a microbiome-driven breeding strategy. These microbial genes were involved in microbial protein synthesis (aroF and serA), carbohydrate metabolism and transport (galT, msmX), lipopolysaccharide biosynthesis (kdsA, lpxD, lpxB), or flagellar synthesis (flgB, fliN) in certain genera within the Proteobacteria phyla (e.g. Serratia, Aeromonas). A microbiome-driven breeding strategy based on these microbial mechanisms as sole information criteria resulted in a positive selection response for both indices (1.36±0.24 and 0.79±0.21 sd of N3 and CLA indices, at 2.06 selection intensity). When evaluating the impact of our microbiome-driven breeding strategy to increase N3 and CLA indices on the environmental trait methane emissions (g/kg of dry matter intake), we obtained a correlated mitigation response of -0.41±0.12 sd.
Conclusion: This research provides insight on the possibility of using the ruminal functional microbiome as information for host genomic selection, which could simultaneously improve several microbiome-driven traits of interest, in this study exemplified with meat quality traits and methane emissions. Video Abstract.
Keywords: Beef; Conjugated linoleic acid; Genomic selection; Methane emissions; Microbial genes; Microbiome-driven breeding; Rumen microbiome; Very long-chain n-3 fatty acids.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Parodi PW. Has the association between saturated fatty acids, serum cholesterol and coronary heart disease been over emphasized? Int Dairy J. 2009;19:345–361. doi: 10.1016/j.idairyj.2009.01.001. - DOI
-
- Parodi PW. Dietary guidelines for saturated fatty acids are not supported by the evidence. Int Dairy J. 2016;52:115–123. doi: 10.1016/j.idairyj.2015.08.007. - DOI
Publication types
MeSH terms
Substances
Grants and funding
- BB/N01720X/1/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/N016742/1/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/S006567/1/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/S006680/1/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Miscellaneous

