Stochastic dynamics and ribosome-RNAP interactions in transcription-translation coupling
- PMID: 36199250
- PMCID: PMC9822797
- DOI: 10.1016/j.bpj.2022.09.041
Stochastic dynamics and ribosome-RNAP interactions in transcription-translation coupling
Abstract
Under certain cellular conditions, transcription and mRNA translation in prokaryotes appear to be "coupled," in which the formation of mRNA transcript and production of its associated protein are temporally correlated. Such transcription-translation coupling (TTC) has been evoked as a mechanism that speeds up the overall process, provides protection against premature termination, and/or regulates the timing of transcript and protein formation. What molecular mechanisms underlie ribosome-RNAP coupling and how they can perform these functions have not been explicitly modeled. We develop and analyze a continuous-time stochastic model that incorporates ribosome and RNAP elongation rates, initiation and termination rates, RNAP pausing, and direct ribosome and RNAP interactions (exclusion and binding). Our model predicts how distributions of delay times depend on these molecular features of transcription and translation. We also propose additional measures for TTC: a direct ribosome-RNAP binding probability and the fraction of time the translation-transcription process is "protected" from attack by transcription-terminating proteins. These metrics quantify different aspects of TTC and differentially depend on parameters of known molecular processes. We use our metrics to reveal how and when our model can exhibit either acceleration or deceleration of transcription, as well as protection from termination. Our detailed mechanistic model provides a basis for designing new experimental assays that can better elucidate the mechanisms of TTC.
Copyright © 2022 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
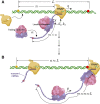

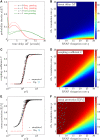


Comment in
-
Transcription-translation coupling: Traveling a road under construction.Biophys J. 2023 Jan 3;122(1):1-3. doi: 10.1016/j.bpj.2022.12.006. Epub 2022 Dec 6. Biophys J. 2023. PMID: 36525978 Free PMC article. No abstract available.
References
-
- Miller O.L., Hamkalo B.A., Thomas C.A. Visualization of bacterial genes in action. Science. 1970;169:392–395. - PubMed
-
- Sydow J.F., Cramer P. RNA polymerase fidelity and transcriptional proofreading. Curr. Opin. Struct. Biol. 2009;19:732–739. Catalysis and regulation/Proteins. - PubMed
-
- Knippa K., Peterson D.O. Fidelity of RNA polymerase II transcription: role of Rbp9 in error detection and proofreading. Biochemistry. 2013;52:7807–7817. - PubMed
-
- Sahoo M., Klumpp S. Backtracking dynamics of RNA polymerase: pausing and error correction. J. Phys. Condens. Matter. 2013;25:374104. - PubMed

