Screening of Sepsis Biomarkers Based on Bioinformatics Data Analysis
- PMID: 36199375
- PMCID: PMC9529510
- DOI: 10.1155/2022/6788569
Screening of Sepsis Biomarkers Based on Bioinformatics Data Analysis
Abstract
Methods: Gene expression profiles of GSE13904, GSE26378, GSE26440, GSE65682, and GSE69528 were obtained from the National Center for Biotechnology Information (NCBI). The differentially expressed genes (DEGs) were searched using limma software package. Gene Ontology (GO) functional analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and protein-protein interaction (PPI) network analysis were performed to elucidate molecular mechanisms of DEGs and screen hub genes.
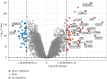
Results: A total of 108 DEGs were identified in the study, of which 67 were upregulated and 41 were downregulated. 15 superlative diagnostic biomarkers (CCL5, CCR7, CD2, CD27, CD274, CD3D, GNLY, GZMA, GZMH, GZMK, IL2RB, IL7R, ITK, KLRB1, and PRF1) for sepsis were identified by bioinformatics analysis.
Conclusion: 15 hub genes (CCL5, CCR7, CD2, CD27, CD274, CD3D, GNLY, GZMA, GZMH, GZMK, IL2RB, IL7R, ITK, KLRB1, and PRF1) have been elucidated in this study, and these biomarkers may be helpful in the diagnosis and therapy of patients with sepsis.
Copyright © 2022 Guibin Liang et al.
Conflict of interest statement
The authors declare that they have not competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

