Expression of vimentin alters cell mechanics, cell-cell adhesion, and gene expression profiles suggesting the induction of a hybrid EMT in human mammary epithelial cells
- PMID: 36200046
- PMCID: PMC9527304
- DOI: 10.3389/fcell.2022.929495
Expression of vimentin alters cell mechanics, cell-cell adhesion, and gene expression profiles suggesting the induction of a hybrid EMT in human mammary epithelial cells
Abstract
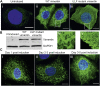
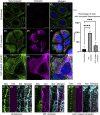
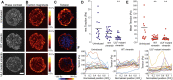
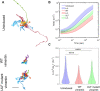
Vimentin is a Type III intermediate filament (VIF) cytoskeletal protein that regulates the mechanical and migratory behavior of cells. Its expression is considered to be a marker for the epithelial to mesenchymal transition (EMT) that takes place in tumor metastasis. However, the molecular mechanisms regulated by the expression of vimentin in the EMT remain largely unexplored. We created MCF7 epithelial cell lines expressing vimentin from a cumate-inducible promoter to address this question. When vimentin expression was induced in these cells, extensive cytoplasmic VIF networks were assembled accompanied by changes in the organization of the endogenous keratin intermediate filament networks and disruption of desmosomes. Significant reductions in intercellular forces by the cells expressing VIFs were measured by quantitative monolayer traction force and stress microscopy. In contrast, laser trapping micro-rheology revealed that the cytoplasm of MCF7 cells expressing VIFs was stiffer than the uninduced cells. Vimentin expression activated transcription of genes involved in pathways responsible for cell migration and locomotion. Importantly, the EMT related transcription factor TWIST1 was upregulated only in wild type vimentin expressing cells and not in cells expressing a mutant non-polymerized form of vimentin, which only formed unit length filaments (ULF). Taken together, our results suggest that vimentin expression induces a hybrid EMT correlated with the upregulation of genes involved in cell migration.
Keywords: Twist1; cell-cell adhesion; desmoplakin; hybrid/partial EMT; intercellular forces; intracellular mechanics; vimentin.
Copyright © 2022 Sivagurunathan, Vahabikashi, Yang, Zhang, Vazquez, Rajasundaram, Politanska, Abdala-Valencia, Notbohm, Guo, Adam and Goldman.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Bar-Kochba E., Toyjanova J., Andrews E., Kim K. S., Franck C. (2015). A fast iterative digital volume correlation algorithm for large deformations. Exp. Mech. 55, 261–274. 10.1007/s11340-014-9874-2 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

