Non-lysine ubiquitylation: Doing things differently
- PMID: 36200073
- PMCID: PMC9527308
- DOI: 10.3389/fmolb.2022.1008175
Non-lysine ubiquitylation: Doing things differently
Abstract
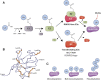
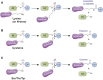
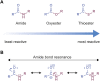
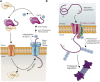
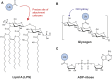
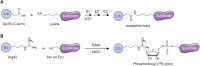
The post-translational modification of proteins with ubiquitin plays a central role in nearly all aspects of eukaryotic biology. Historically, studies have focused on the conjugation of ubiquitin to lysine residues in substrates, but it is now clear that ubiquitylation can also occur on cysteine, serine, and threonine residues, as well as on the N-terminal amino group of proteins. Paradigm-shifting reports of non-proteinaceous substrates have further extended the reach of ubiquitylation beyond the proteome to include intracellular lipids and sugars. Additionally, results from bacteria have revealed novel ways to ubiquitylate (and deubiquitylate) substrates without the need for any of the enzymatic components of the canonical ubiquitylation cascade. Focusing mainly upon recent findings, this review aims to outline the current understanding of non-lysine ubiquitylation and speculate upon the molecular mechanisms and physiological importance of this non-canonical modification.
Keywords: ERAD; LUBAC; Rnf213; non-canonical ubiquitylation; non-lysine ubiquitylation; oxyester bond; thioester bond; ubiquitin.
Copyright © 2022 Kelsall.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Ahel J., Fletcher A., Grabarczyk D. B., Roitinger E., Deszcz L., Lehner A., et al. (2021). E3 ubiquitin ligase RNF213 employs a non-canonical zinc finger active site and is allosterically regulated by ATP. bioRxiv 2021, 443411. 10.1101/2021.05.10.443411 - DOI
Publication types
LinkOut - more resources
Full Text Sources

