Novel NRF2-activated cancer treatments utilizing synthetic lethality
- PMID: 36200139
- PMCID: PMC10092255
- DOI: 10.1002/iub.2680
Novel NRF2-activated cancer treatments utilizing synthetic lethality
Abstract
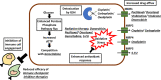
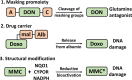
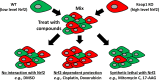
The KEAP1-NRF2 pathway regulates the main inducible cellular response to oxidative and electrophilic stresses. Activating mutations in the KEAP1-NRF2 pathway occur commonly in human cancer, where they contribute to the formation of aggressive tumours that are associated with a poor prognosis for patients. An important clinical feature of these tumours is their defiance to all current anti-cancer treatment regimens, highlighting the need for the development of new therapeutic strategies to target NRF2-activated cancers. In this review, we discuss the mechanisms through which acquired NRF2 hyperactivation can result in resistance of tumours to immune checkpoint inhibitor therapies in addition to classical chemotherapeutics, and propose with examples that using a synthetic lethal strategy mediated by NRF2-target gene-dependent bioactivation of prodrugs represents a promising strategy to specifically enhance toxicity to heretofore untreatable NRF2-hyperactivated human tumours.
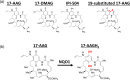
Keywords: KEAP1; NFE2L2; NRF2; bioactivation; cancer; mitomycin C; oxidative stress; prodrug; stress response; synthetic lethal.
© 2022 The Authors. IUBMB Life published by Wiley Periodicals LLC on behalf of International Union of Biochemistry and Molecular Biology.
Figures






Similar articles
-
NRF2-Dependent Bioactivation of Mitomycin C as a Novel Strategy To Target KEAP1-NRF2 Pathway Activation in Human Cancer.Mol Cell Biol. 2021 Jan 25;41(2):e00473-20. doi: 10.1128/MCB.00473-20. Print 2021 Jan 25. Mol Cell Biol. 2021. PMID: 33139492 Free PMC article.
-
Immunoediting of KEAP1-NRF2 mutant tumours is required to circumvent NRF2-mediated immune surveillance.Redox Biol. 2023 Nov;67:102904. doi: 10.1016/j.redox.2023.102904. Epub 2023 Oct 10. Redox Biol. 2023. PMID: 37839356 Free PMC article.
-
Molecular Basis of the KEAP1-NRF2 Signaling Pathway.Mol Cells. 2023 Mar 31;46(3):133-141. doi: 10.14348/molcells.2023.0028. Epub 2023 Mar 27. Mol Cells. 2023. PMID: 36994473 Free PMC article. Review.
-
Geldanamycin-Derived HSP90 Inhibitors Are Synthetic Lethal with NRF2.Mol Cell Biol. 2020 Oct 26;40(22):e00377-20. doi: 10.1128/MCB.00377-20. Print 2020 Oct 26. Mol Cell Biol. 2020. PMID: 32868290 Free PMC article.
-
[Effect of Nrf2 in tumor progression and its inhibitors in cancer therapy].Zhongguo Zhong Yao Za Zhi. 2021 Jan;46(1):24-32. doi: 10.19540/j.cnki.cjcmm.20200915.601. Zhongguo Zhong Yao Za Zhi. 2021. PMID: 33645047 Review. Chinese.
Cited by
-
Geldanamycin, a Naturally Occurring Inhibitor of Hsp90 and a Lead Compound for Medicinal Chemistry.J Med Chem. 2024 Oct 24;67(20):17946-17963. doi: 10.1021/acs.jmedchem.4c01048. Epub 2024 Oct 3. J Med Chem. 2024. PMID: 39361055 Free PMC article. Review.
-
Lymphatic Metastasis of Esophageal Squamous Cell Carcinoma: The Role of NRF2 and Therapeutic Strategies.Cancers (Basel). 2025 May 31;17(11):1853. doi: 10.3390/cancers17111853. Cancers (Basel). 2025. PMID: 40507333 Free PMC article. Review.
-
Nrf2 in TIME: The Emerging Role of Nuclear Factor Erythroid 2-Related Factor 2 in the Tumor Immune Microenvironment.Mol Cells. 2023 Mar 31;46(3):142-152. doi: 10.14348/molcells.2023.2183. Epub 2023 Mar 17. Mol Cells. 2023. PMID: 36927604 Free PMC article. Review.
-
Redox Signaling Modulates Activity of Immune Checkpoint Inhibitors in Cancer Patients.Biomedicines. 2023 Apr 29;11(5):1325. doi: 10.3390/biomedicines11051325. Biomedicines. 2023. PMID: 37238995 Free PMC article. Review.
-
Acyl Urea Compounds Therapeutics and its Inhibition for Cancers in Women: A Review.Anticancer Agents Med Chem. 2025;25(2):86-98. doi: 10.2174/0118715206330232240913100744. Anticancer Agents Med Chem. 2025. PMID: 39318218 Review.
References
-
- Itoh K, Chiba T, Takahashi S, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236(2):313–322. - PubMed
-
- McMahon M, Itoh K, Yamamoto M, et al. The Cap'n'Collar basic leucine zipper transcription factor Nrf2 (NF‐E2 p45‐related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61(8):3299–3307. - PubMed
-
- Kwak M‐K, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1‐Nrf2 pathway. Identification of novel gene clusters for cell survival. J Biol Chem. 2003;278:8135–8145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

