rRNA expansion segment 7 in eukaryotes: from Signature Fold to tentacles
- PMID: 36200812
- PMCID: PMC9561286
- DOI: 10.1093/nar/gkac844
rRNA expansion segment 7 in eukaryotes: from Signature Fold to tentacles
Abstract
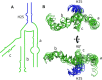
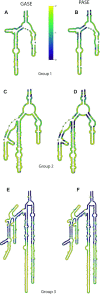
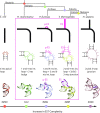
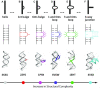
The ribosomal core is universally conserved across the tree of life. However, eukaryotic ribosomes contain diverse rRNA expansion segments (ESs) on their surfaces. Sites of ES insertions are predicted from sites of insertion of micro-ESs in archaea. Expansion segment 7 (ES7) is one of the most diverse regions of the ribosome, emanating from a short stem loop and ranging to over 750 nucleotides in mammals. We present secondary and full-atom 3D structures of ES7 from species spanning eukaryotic diversity. Our results are based on experimental 3D structures, the accretion model of ribosomal evolution, phylogenetic relationships, multiple sequence alignments, RNA folding algorithms and 3D modeling by RNAComposer. ES7 contains a distinct motif, the 'ES7 Signature Fold', which is generally invariant in 2D topology and 3D structure in all eukaryotic ribosomes. We establish a model in which ES7 developed over evolution through a series of elementary and recursive growth events. The data are sufficient to support an atomic-level accretion path for rRNA growth. The non-monophyletic distribution of some ES7 features across the phylogeny suggests acquisition via convergent processes. And finally, illustrating the power of our approach, we constructed the 2D and 3D structure of the entire LSU rRNA of Mus musculus.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Yeast rRNA Expansion Segments: Folding and Function.J Mol Biol. 2016 Oct 9;428(20):4048-4059. doi: 10.1016/j.jmb.2016.08.008. Epub 2016 Aug 10. J Mol Biol. 2016. PMID: 27521697
-
Supersized Ribosomal RNA Expansion Segments in Asgard Archaea.Genome Biol Evol. 2020 Oct 1;12(10):1694-1710. doi: 10.1093/gbe/evaa170. Genome Biol Evol. 2020. PMID: 32785681 Free PMC article.
-
G-Quadruplexes in Human Ribosomal RNA.J Mol Biol. 2019 May 3;431(10):1940-1955. doi: 10.1016/j.jmb.2019.03.010. Epub 2019 Mar 15. J Mol Biol. 2019. PMID: 30885721 Free PMC article.
-
The story of rRNA expansion segments: Finding functionality amidst diversity.Wiley Interdiscip Rev RNA. 2023 Jan;14(1):e1732. doi: 10.1002/wrna.1732. Epub 2022 Apr 15. Wiley Interdiscip Rev RNA. 2023. PMID: 35429135 Review.
-
Pseudouridines and pseudouridine synthases of the ribosome.Cold Spring Harb Symp Quant Biol. 2001;66:147-59. doi: 10.1101/sqb.2001.66.147. Cold Spring Harb Symp Quant Biol. 2001. PMID: 12762017 Review.
Cited by
-
TRMT1L-catalyzed m22G27 on tyrosine tRNA is required for efficient mRNA translation and cell survival under oxidative stress.Cell Rep. 2025 Jan 28;44(1):115167. doi: 10.1016/j.celrep.2024.115167. Epub 2025 Jan 8. Cell Rep. 2025. PMID: 39786998 Free PMC article.
-
Structures of Naked Mole-Rat, Tuco-Tuco, and Guinea Pig Ribosomes-Is rRNA Fragmentation Linked to Translational Fidelity?bioRxiv [Preprint]. 2025 Aug 2:2025.08.01.667930. doi: 10.1101/2025.08.01.667930. bioRxiv. 2025. PMID: 40766631 Free PMC article. Preprint.
-
bpRNA-CosMoS: a robust and efficient RNA structural comparison method using k-mer based cosine similarity.Bioinformatics. 2025 Mar 29;41(4):btaf108. doi: 10.1093/bioinformatics/btaf108. Bioinformatics. 2025. PMID: 40085007 Free PMC article.
-
TRMT1L-catalyzed m2 2G27 on tyrosine tRNA is required for efficient mRNA translation and cell survival under oxidative stress.bioRxiv [Preprint]. 2024 Oct 12:2024.05.02.591343. doi: 10.1101/2024.05.02.591343. bioRxiv. 2024. Update in: Cell Rep. 2025 Jan 28;44(1):115167. doi: 10.1016/j.celrep.2024.115167. PMID: 39416027 Free PMC article. Updated. Preprint.
-
Reduction of Ribosomal Expansion Segments in Yeast Species of the Magnusiomyces/Saprochaete Clade.Genome Biol Evol. 2024 Aug 5;16(8):evae173. doi: 10.1093/gbe/evae173. Genome Biol Evol. 2024. PMID: 39119893 Free PMC article.
References
-
- Melnikov S., Ben-Shem A., Garreau de Loubresse N., Jenner L., Yusupova G., Yusupov M.. One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 2012; 19:560–567. - PubMed
-
- Bowman J.C., Petrov A.S., Frenkel-Pinter M., Penev P.I., Williams L.D.. Root of the tree: the significance, evolution, and origins of the ribosome. Chem. Rev. 2020; 120:4848–4878. - PubMed
-
- Anger A.M., Armache J.P., Berninghausen O., Habeck M., Subklewe M., Wilson D.N., Beckmann R.. Structures of the human and drosophila 80S ribosome. Nature. 2013; 497:80–85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

